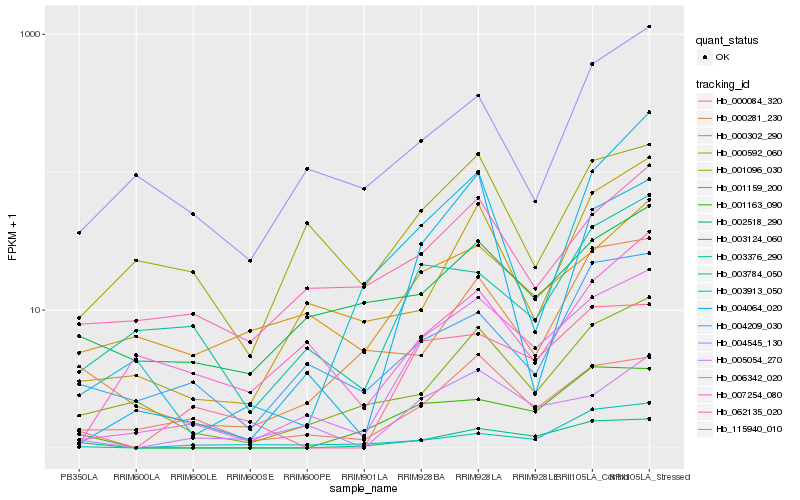
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006342_020 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_005054_270 |
0.1638622996 |
- |
- |
- |
| 3 |
Hb_115940_010 |
0.1760981589 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |
| 4 |
Hb_000084_320 |
0.1854729597 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 5 |
Hb_001159_200 |
0.1890259816 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000592_060 |
0.1979162847 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 7 |
Hb_003376_290 |
0.2022226977 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 8 |
Hb_003124_060 |
0.2059993096 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 9 |
Hb_000281_230 |
0.2135295905 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 10 |
Hb_062135_020 |
0.2141282129 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 11 |
Hb_004209_030 |
0.2171026311 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000302_290 |
0.2178635005 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 13 |
Hb_004064_020 |
0.2182217448 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 14 |
Hb_007254_080 |
0.2205003217 |
- |
- |
- |
| 15 |
Hb_003784_050 |
0.2205255511 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 16 |
Hb_001163_090 |
0.2214087049 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 17 |
Hb_003913_050 |
0.2224767922 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 18 |
Hb_001096_030 |
0.2229997471 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 19 |
Hb_004545_130 |
0.2231746789 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 20 |
Hb_002518_290 |
0.224037564 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |