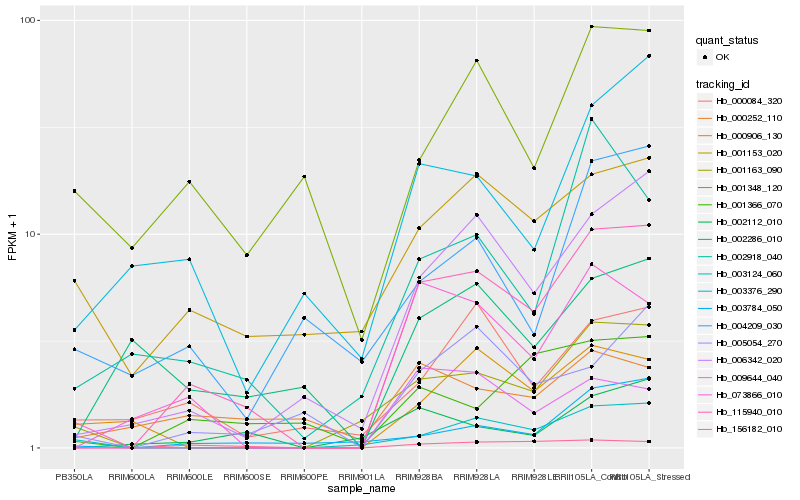
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115940_010 |
0.0 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |
| 2 |
Hb_006342_020 |
0.1760981589 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_073866_010 |
0.2002427815 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 4 |
Hb_156182_010 |
0.2006272603 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 5 |
Hb_001163_090 |
0.2141085742 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 6 |
Hb_003124_060 |
0.2231763253 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 7 |
Hb_005054_270 |
0.2233669034 |
- |
- |
- |
| 8 |
Hb_000084_320 |
0.2325551303 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 9 |
Hb_001366_070 |
0.2367144596 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001153_020 |
0.236963002 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 11 |
Hb_002918_040 |
0.2384499218 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 12 |
Hb_003784_050 |
0.239381168 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 13 |
Hb_003376_290 |
0.2404072412 |
- |
- |
hypothetical protein POPTR_0010s22880g [Populus trichocarpa] |
| 14 |
Hb_009644_040 |
0.2428066811 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 15 |
Hb_002112_010 |
0.2430048021 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 16 |
Hb_001348_120 |
0.257102421 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |
| 17 |
Hb_000906_130 |
0.2612901376 |
- |
- |
- |
| 18 |
Hb_002286_010 |
0.2615984575 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 19 |
Hb_000252_110 |
0.2636253855 |
- |
- |
- |
| 20 |
Hb_004209_030 |
0.2644010514 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |