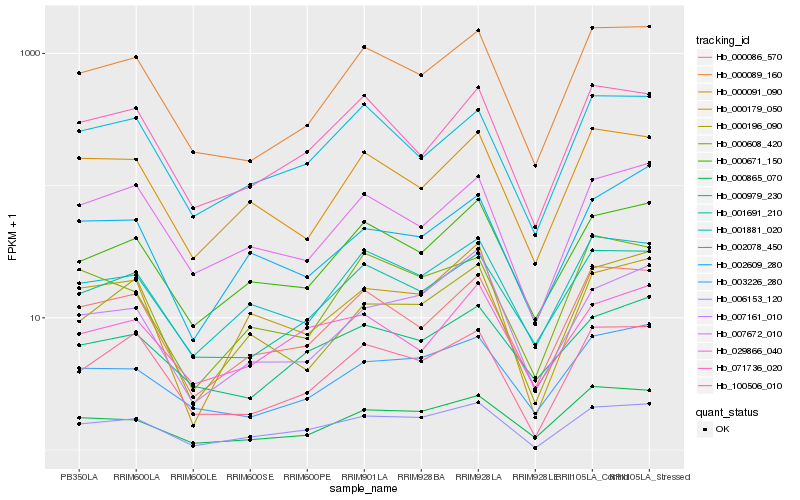
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 2 |
Hb_000179_050 |
0.0966804655 |
- |
- |
40S ribosomal protein S17A [Hevea brasiliensis] |
| 3 |
Hb_000671_150 |
0.1003659722 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 4 |
Hb_000086_570 |
0.1007164158 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 5 |
Hb_100506_010 |
0.1008545333 |
- |
- |
PREDICTED: uncharacterized protein LOC105133651 isoform X2 [Populus euphratica] |
| 6 |
Hb_001881_020 |
0.1039650719 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 7 |
Hb_000089_160 |
0.1047670555 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 8 |
Hb_001691_210 |
0.111230007 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 9 |
Hb_007672_010 |
0.1144541841 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002078_450 |
0.116025062 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 11 |
Hb_003226_280 |
0.1160499689 |
- |
- |
PREDICTED: peter Pan-like protein [Populus euphratica] |
| 12 |
Hb_071736_020 |
0.1165156881 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 13 |
Hb_007161_010 |
0.1172320129 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000608_420 |
0.1204010741 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 15 |
Hb_000196_090 |
0.1220680319 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 16 |
Hb_000865_070 |
0.1233424906 |
- |
- |
Protein FAR1-RELATED SEQUENCE 3 [Glycine soja] |
| 17 |
Hb_000091_090 |
0.1233966455 |
- |
- |
hypothetical protein JCGZ_09031 [Jatropha curcas] |
| 18 |
Hb_029866_040 |
0.1246101412 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 19 |
Hb_000979_230 |
0.1259597647 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |
| 20 |
Hb_002609_280 |
0.1277851665 |
- |
- |
villin 1-4, putative [Ricinus communis] |