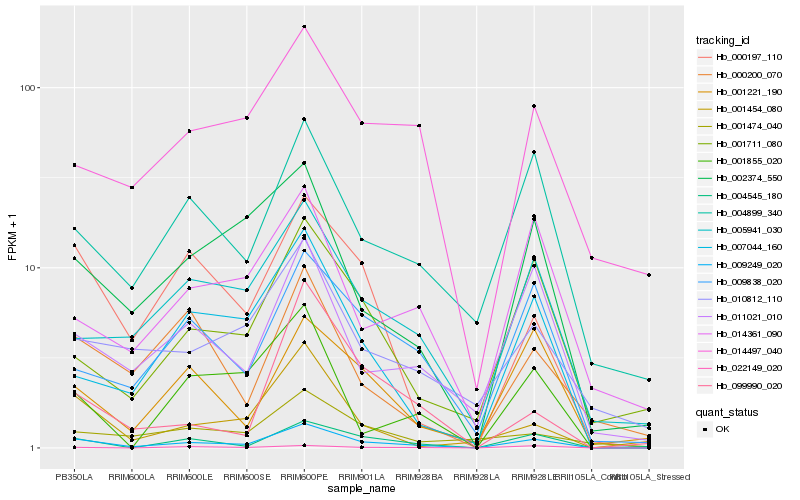
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009249_020 |
0.0 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 2 |
Hb_004545_180 |
0.1829460564 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_009838_020 |
0.203385116 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 4 |
Hb_011021_010 |
0.2042693853 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000197_110 |
0.2043235337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004899_340 |
0.2055532543 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_007044_160 |
0.2060877132 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 8 |
Hb_005941_030 |
0.2063810063 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002374_550 |
0.2090698226 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 10 |
Hb_001711_080 |
0.2146136016 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 11 |
Hb_001221_190 |
0.2150195531 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 12 |
Hb_001454_080 |
0.2182837418 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_014497_040 |
0.2263870641 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010812_110 |
0.2268334503 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 15 |
Hb_001855_020 |
0.2283954147 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_022149_020 |
0.2286427062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_014361_090 |
0.2345307893 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 18 |
Hb_000200_070 |
0.2377973498 |
- |
- |
PREDICTED: CDP-diacylglycerol--serine O-phosphatidyltransferase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001474_040 |
0.2458045203 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 20 |
Hb_099990_020 |
0.2470992608 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |