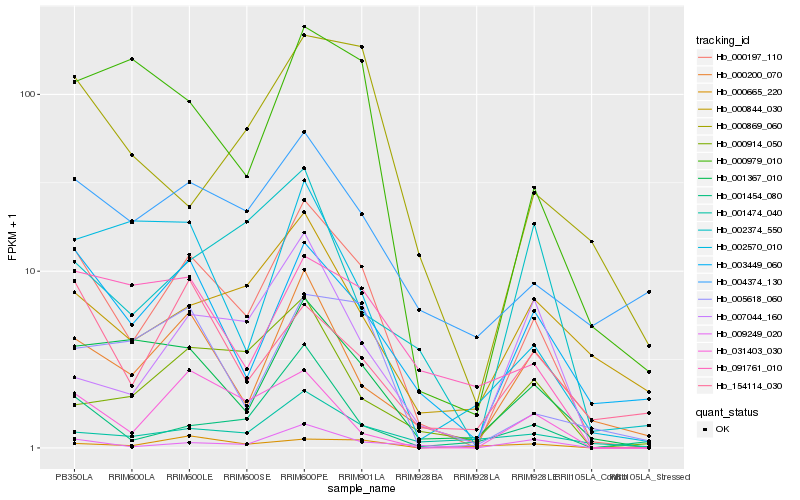
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003449_060 |
0.1985491286 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 3 |
Hb_001367_010 |
0.2036101628 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 4 |
Hb_009249_020 |
0.2043235337 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 5 |
Hb_000200_070 |
0.2047312051 |
- |
- |
PREDICTED: CDP-diacylglycerol--serine O-phosphatidyltransferase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004374_130 |
0.2259358684 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 7 |
Hb_031403_030 |
0.2287469543 |
- |
- |
- |
| 8 |
Hb_000979_010 |
0.2340479241 |
- |
- |
PREDICTED: uncharacterized protein LOC105128288 [Populus euphratica] |
| 9 |
Hb_005618_060 |
0.2409660116 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 10 |
Hb_091761_010 |
0.2417676804 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 11 |
Hb_001474_040 |
0.2425381546 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 12 |
Hb_001454_080 |
0.2443786554 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 13 |
Hb_000844_030 |
0.2462357257 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 14 |
Hb_154114_030 |
0.2489299249 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_002570_010 |
0.2527491321 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 16 |
Hb_000869_060 |
0.2585334014 |
- |
- |
hypothetical protein L484_003245 [Morus notabilis] |
| 17 |
Hb_000914_050 |
0.2589859658 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 18 |
Hb_007044_160 |
0.259207496 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 19 |
Hb_000665_220 |
0.2645851923 |
- |
- |
catalytic, putative [Ricinus communis] |
| 20 |
Hb_002374_550 |
0.270024999 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |