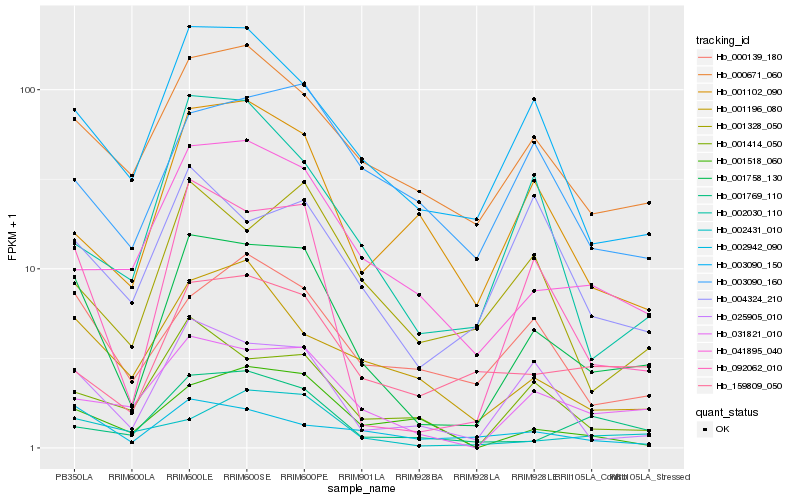
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001758_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 2 |
Hb_092062_010 |
0.1463108006 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 3 |
Hb_031821_010 |
0.1672235759 |
- |
- |
ccd1, putative [Ricinus communis] |
| 4 |
Hb_025905_010 |
0.1703636846 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 5 |
Hb_003090_150 |
0.1808756518 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 6 |
Hb_000671_060 |
0.1845252154 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001414_050 |
0.196312969 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 8 |
Hb_041895_040 |
0.202011376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000139_180 |
0.2032226581 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 10 |
Hb_159809_050 |
0.2089133705 |
- |
- |
AT14A, putative [Ricinus communis] |
| 11 |
Hb_001328_050 |
0.2092797303 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 12 |
Hb_002431_010 |
0.212199118 |
- |
- |
PREDICTED: uncharacterized protein LOC104599557 isoform X2 [Nelumbo nucifera] |
| 13 |
Hb_001196_080 |
0.2122629628 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 14 |
Hb_002030_110 |
0.2176993813 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 15 |
Hb_001769_110 |
0.2179197608 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 16 |
Hb_001102_090 |
0.2182935859 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 17 |
Hb_002942_090 |
0.2199066389 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 18 |
Hb_004324_210 |
0.2199516647 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_001518_060 |
0.2199597693 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 20 |
Hb_003090_160 |
0.2221668329 |
- |
- |
- |