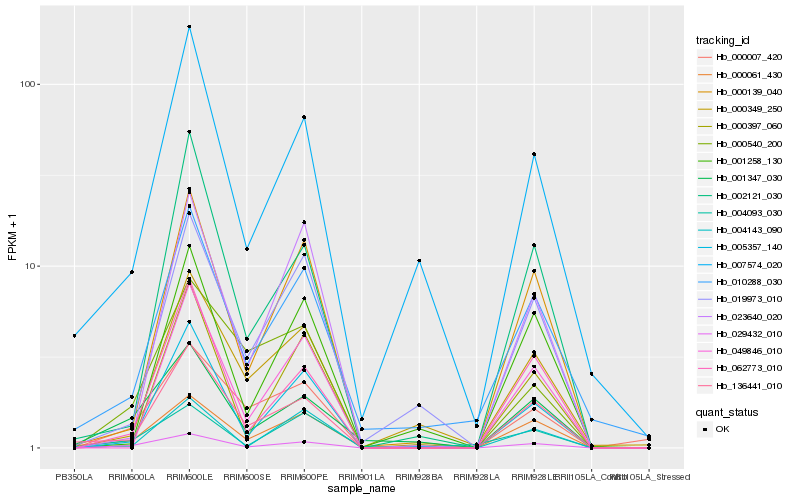
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029432_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001347_030 |
0.1477923576 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000061_430 |
0.1486521739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_136441_010 |
0.1612175615 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 5 |
Hb_005357_140 |
0.1640678095 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein A precursor, putative [Ricinus communis] |
| 6 |
Hb_062773_010 |
0.1642117356 |
- |
- |
beta glucosidase [Manihot esculenta] |
| 7 |
Hb_000139_040 |
0.1790412879 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 8 |
Hb_000007_420 |
0.1797070094 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 9 |
Hb_010288_030 |
0.1901269302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 10 |
Hb_007574_020 |
0.1945646825 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 11 |
Hb_004093_030 |
0.1991015428 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 12 |
Hb_019973_010 |
0.1992457541 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 13 |
Hb_049846_010 |
0.2058166979 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 14 |
Hb_002121_030 |
0.2065128623 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 15 |
Hb_000540_200 |
0.2085155467 |
- |
- |
Uncharacterized protein TCM_041994 [Theobroma cacao] |
| 16 |
Hb_004143_090 |
0.2089777825 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_023640_020 |
0.2104849239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000397_060 |
0.2107671184 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 19 |
Hb_001258_130 |
0.2108424635 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 20 |
Hb_000349_250 |
0.2108437872 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |