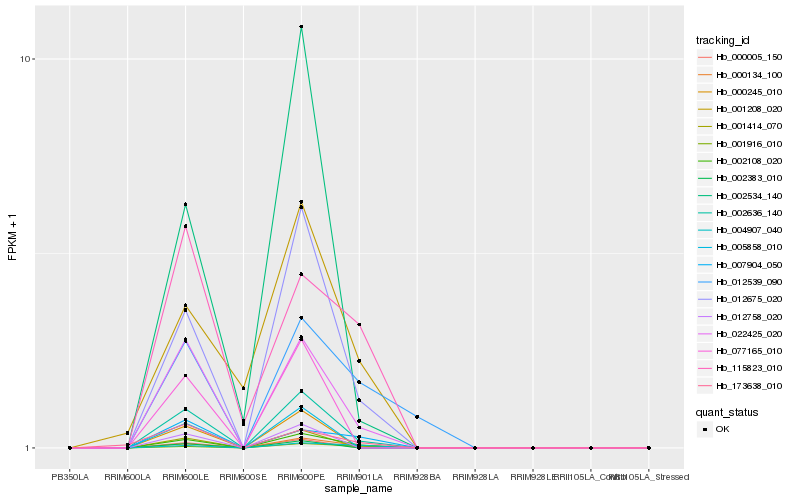
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002108_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_16012 [Jatropha curcas] |
| 2 |
Hb_000134_100 |
0.0725170584 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 3 |
Hb_002636_140 |
0.081854559 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691-like [Populus euphratica] |
| 4 |
Hb_012675_020 |
0.0921375828 |
- |
- |
hypothetical protein CISIN_1g018227mg [Citrus sinensis] |
| 5 |
Hb_022425_020 |
0.1242005091 |
- |
- |
- |
| 6 |
Hb_001916_010 |
0.165403211 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 7 |
Hb_004907_040 |
0.1732558823 |
- |
- |
hypothetical protein POPTR_0011s079751g, partial [Populus trichocarpa] |
| 8 |
Hb_007904_050 |
0.2177021719 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 9 |
Hb_012539_090 |
0.2228406491 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 10 |
Hb_001208_020 |
0.2249076407 |
- |
- |
- |
| 11 |
Hb_002534_140 |
0.2367020692 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 12 |
Hb_173638_010 |
0.2368841286 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_115823_010 |
0.2391254894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005858_010 |
0.2392523512 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_12260 [Jatropha curcas] |
| 15 |
Hb_012758_020 |
0.2392537168 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_002383_010 |
0.2393099743 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 17 |
Hb_000005_150 |
0.2393340148 |
- |
- |
PREDICTED: cation/H(+) antiporter 10-like [Jatropha curcas] |
| 18 |
Hb_000245_010 |
0.2393572554 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Populus euphratica] |
| 19 |
Hb_077165_010 |
0.2393576034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001414_070 |
0.2395694496 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |