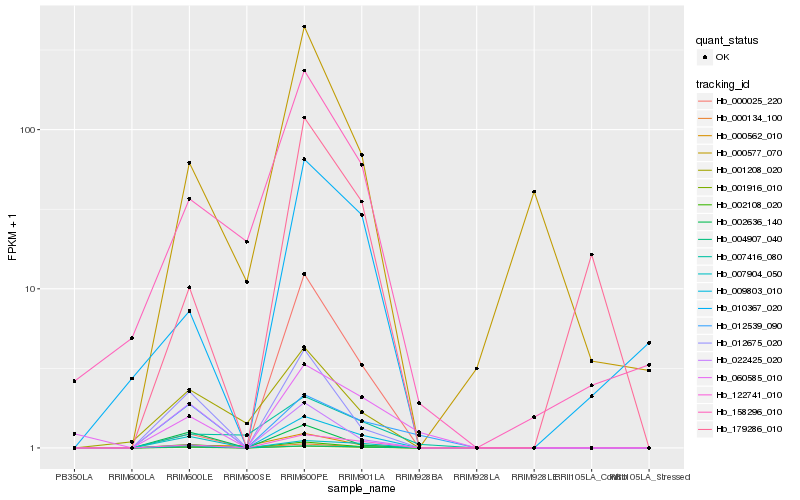
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001916_010 |
0.0 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 2 |
Hb_004907_040 |
0.0688956312 |
- |
- |
hypothetical protein POPTR_0011s079751g, partial [Populus trichocarpa] |
| 3 |
Hb_122741_010 |
0.082464374 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |
| 4 |
Hb_000134_100 |
0.1027605075 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 5 |
Hb_002108_020 |
0.165403211 |
- |
- |
hypothetical protein JCGZ_16012 [Jatropha curcas] |
| 6 |
Hb_012675_020 |
0.1964710494 |
- |
- |
hypothetical protein CISIN_1g018227mg [Citrus sinensis] |
| 7 |
Hb_009803_010 |
0.2027075906 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 8 |
Hb_158296_010 |
0.2393516714 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 9 |
Hb_002636_140 |
0.2419592362 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691-like [Populus euphratica] |
| 10 |
Hb_001208_020 |
0.2425216265 |
- |
- |
- |
| 11 |
Hb_010367_020 |
0.242725802 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 12 |
Hb_060585_010 |
0.2429139111 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_012539_090 |
0.245907375 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 14 |
Hb_000562_010 |
0.2612262501 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 15 |
Hb_007416_080 |
0.2684047354 |
- |
- |
- |
| 16 |
Hb_000577_070 |
0.2764659222 |
- |
- |
- |
| 17 |
Hb_179286_010 |
0.2789489539 |
- |
- |
STRUWWELPETER family protein [Populus trichocarpa] |
| 18 |
Hb_022425_020 |
0.2799515683 |
- |
- |
- |
| 19 |
Hb_007904_050 |
0.2850475582 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 20 |
Hb_000025_220 |
0.2852967877 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |