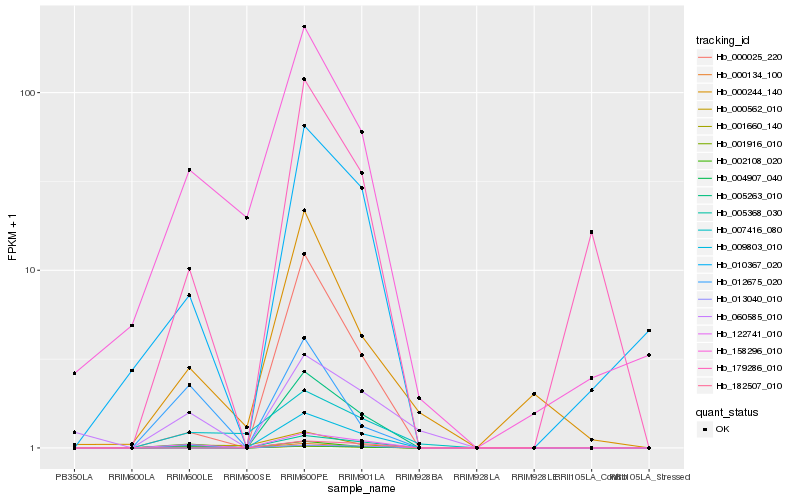
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122741_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |
| 2 |
Hb_001916_010 |
0.082464374 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 3 |
Hb_004907_040 |
0.1231081629 |
- |
- |
hypothetical protein POPTR_0011s079751g, partial [Populus trichocarpa] |
| 4 |
Hb_009803_010 |
0.1386599085 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 5 |
Hb_000134_100 |
0.1781830911 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Prunus mume] |
| 6 |
Hb_010367_020 |
0.1996685765 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 7 |
Hb_158296_010 |
0.2323537532 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 8 |
Hb_060585_010 |
0.2363281434 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000025_220 |
0.2410955669 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 10 |
Hb_002108_020 |
0.2442290139 |
- |
- |
hypothetical protein JCGZ_16012 [Jatropha curcas] |
| 11 |
Hb_179286_010 |
0.2495896927 |
- |
- |
STRUWWELPETER family protein [Populus trichocarpa] |
| 12 |
Hb_007416_080 |
0.2570982212 |
- |
- |
- |
| 13 |
Hb_005368_030 |
0.2584396528 |
- |
- |
- |
| 14 |
Hb_012675_020 |
0.2613604761 |
- |
- |
hypothetical protein CISIN_1g018227mg [Citrus sinensis] |
| 15 |
Hb_000562_010 |
0.2632630742 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 16 |
Hb_182507_010 |
0.2642085027 |
- |
- |
- |
| 17 |
Hb_001660_140 |
0.2649012527 |
- |
- |
- |
| 18 |
Hb_005263_010 |
0.2670715879 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 19 |
Hb_013040_010 |
0.2703231936 |
- |
- |
hypothetical protein POPTR_0009s09690g [Populus trichocarpa] |
| 20 |
Hb_000244_140 |
0.2747255743 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |