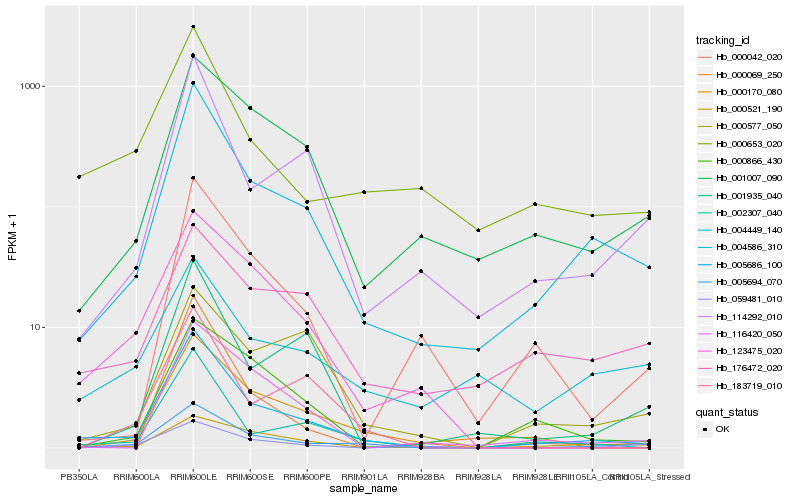
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_070 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_005686_100 |
0.1556168922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004586_310 |
0.1713294819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001007_090 |
0.2316293161 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 5 |
Hb_000521_190 |
0.2358905699 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 6 |
Hb_123475_020 |
0.2362477656 |
- |
- |
- |
| 7 |
Hb_059481_010 |
0.2370263884 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 8 |
Hb_000653_020 |
0.2386753024 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 9 |
Hb_114292_010 |
0.239381771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004449_140 |
0.2416312309 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 11 |
Hb_000170_080 |
0.2501431889 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 12 |
Hb_002307_040 |
0.2530811397 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_176472_020 |
0.2573820784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_183719_010 |
0.2577791347 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 15 |
Hb_000866_430 |
0.2611705503 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 16 |
Hb_116420_050 |
0.2619324187 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 17 |
Hb_000069_250 |
0.2662575455 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 18 |
Hb_001935_040 |
0.2680473292 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 19 |
Hb_000577_050 |
0.268892017 |
- |
- |
- |
| 20 |
Hb_000042_020 |
0.2714124551 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |