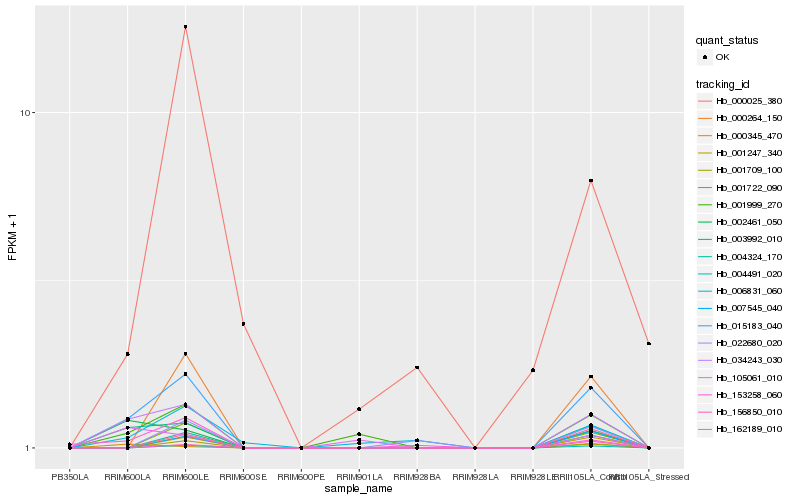
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015183_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_034243_030 |
0.1098922061 |
- |
- |
B3 domain-containing protein [Medicago truncatula] |
| 3 |
Hb_003992_010 |
0.2050512679 |
- |
- |
- |
| 4 |
Hb_001999_270 |
0.2529304981 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 5 |
Hb_002461_050 |
0.2734969605 |
- |
- |
- |
| 6 |
Hb_004491_020 |
0.2904931661 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 7 |
Hb_000264_150 |
0.2910041101 |
- |
- |
PREDICTED: uncharacterized protein LOC103870759 [Brassica rapa] |
| 8 |
Hb_001722_090 |
0.2945925461 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 9 |
Hb_001247_340 |
0.2946438048 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_001709_100 |
0.2947340428 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 11 |
Hb_153258_060 |
0.3125476207 |
- |
- |
- |
| 12 |
Hb_162189_010 |
0.3149398947 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 13 |
Hb_007545_040 |
0.3171947367 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_006831_060 |
0.3189378492 |
- |
- |
hypothetical protein JCGZ_16419 [Jatropha curcas] |
| 15 |
Hb_004324_170 |
0.3200056385 |
- |
- |
- |
| 16 |
Hb_000345_470 |
0.3339881163 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_156850_010 |
0.3446072992 |
- |
- |
PREDICTED: uncharacterized protein LOC105635403 [Jatropha curcas] |
| 18 |
Hb_000025_380 |
0.367686276 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 19 |
Hb_022680_020 |
0.3766523194 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 20 |
Hb_105061_010 |
0.3848646534 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |