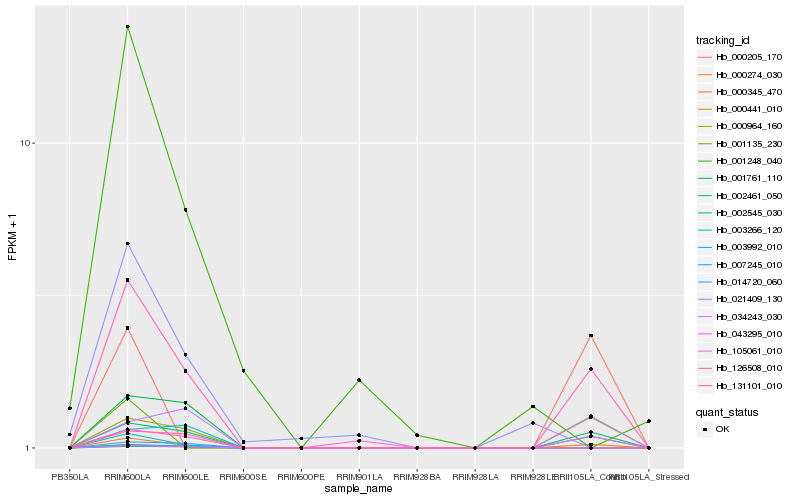
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126508_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002461_050 |
0.1427020918 |
- |
- |
- |
| 3 |
Hb_000345_470 |
0.2161654395 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 4 |
Hb_003992_010 |
0.2507769771 |
- |
- |
- |
| 5 |
Hb_105061_010 |
0.3053262645 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 6 |
Hb_034243_030 |
0.3075271075 |
- |
- |
B3 domain-containing protein [Medicago truncatula] |
| 7 |
Hb_000274_030 |
0.3276663077 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 8 |
Hb_003266_120 |
0.332826604 |
- |
- |
PREDICTED: uncharacterized protein LOC105650228 [Jatropha curcas] |
| 9 |
Hb_001135_230 |
0.3394225422 |
- |
- |
PREDICTED: protein REVEILLE 6-like isoform X3 [Gossypium raimondii] |
| 10 |
Hb_043295_010 |
0.3469584673 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 11 |
Hb_007245_010 |
0.346991271 |
- |
- |
PREDICTED: uncharacterized protein LOC104783698 [Camelina sativa] |
| 12 |
Hb_000964_160 |
0.3481188758 |
- |
- |
PREDICTED: uncharacterized protein LOC102617979 isoform X2 [Citrus sinensis] |
| 13 |
Hb_002545_030 |
0.363639132 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000441_010 |
0.365708918 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 15 |
Hb_014720_060 |
0.3660169261 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 16 |
Hb_131101_010 |
0.3727417409 |
- |
- |
- |
| 17 |
Hb_001761_110 |
0.3747394346 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 18 |
Hb_000205_170 |
0.3751090521 |
- |
- |
PREDICTED: uncharacterized protein LOC103926801 [Pyrus x bretschneideri] |
| 19 |
Hb_001248_040 |
0.3805079035 |
- |
- |
- |
| 20 |
Hb_021409_130 |
0.3893128324 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like isoform 1 [Glycine max] |