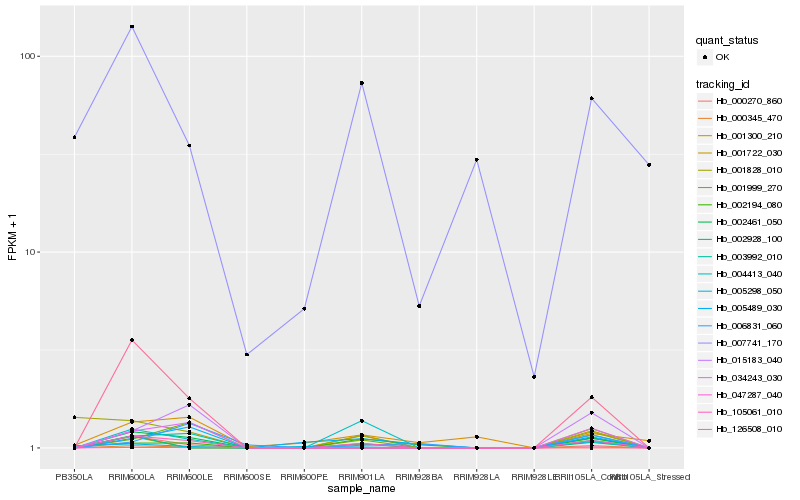
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105061_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_001999_270 |
0.2718445204 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 3 |
Hb_004413_040 |
0.2720210308 |
- |
- |
- |
| 4 |
Hb_002461_050 |
0.275252622 |
- |
- |
- |
| 5 |
Hb_126508_010 |
0.3053262645 |
- |
- |
- |
| 6 |
Hb_003992_010 |
0.3093852519 |
- |
- |
- |
| 7 |
Hb_000345_470 |
0.3154845926 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 8 |
Hb_034243_030 |
0.3231303027 |
- |
- |
B3 domain-containing protein [Medicago truncatula] |
| 9 |
Hb_047287_040 |
0.35211697 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 10 |
Hb_005489_030 |
0.3541983774 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 11 |
Hb_002928_100 |
0.3612068701 |
- |
- |
- |
| 12 |
Hb_002194_080 |
0.3765833864 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 13 |
Hb_015183_040 |
0.3848646534 |
- |
- |
- |
| 14 |
Hb_000270_860 |
0.3850351503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001300_210 |
0.4008735232 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 16 |
Hb_001722_030 |
0.4031924184 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105123768 isoform X2 [Populus euphratica] |
| 17 |
Hb_007741_170 |
0.4052481868 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4-like [Jatropha curcas] |
| 18 |
Hb_005298_050 |
0.4113022464 |
- |
- |
Integrase core domain, putative [Oryza sativa Japonica Group] |
| 19 |
Hb_001828_010 |
0.4216214878 |
- |
- |
- |
| 20 |
Hb_006831_060 |
0.4225200816 |
- |
- |
hypothetical protein JCGZ_16419 [Jatropha curcas] |