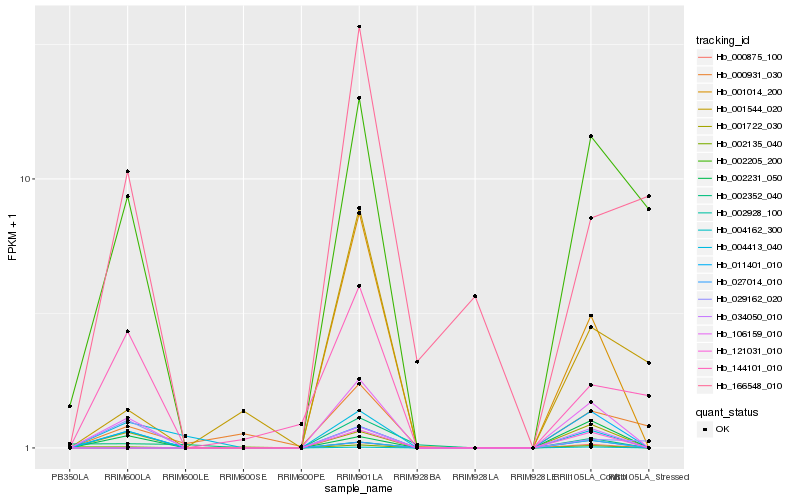
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106159_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001722_030 |
0.1874804421 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105123768 isoform X2 [Populus euphratica] |
| 3 |
Hb_011401_010 |
0.2074619595 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 4 |
Hb_002231_050 |
0.2699430196 |
- |
- |
- |
| 5 |
Hb_004413_040 |
0.2841541345 |
- |
- |
- |
| 6 |
Hb_002205_200 |
0.2846903397 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 7 |
Hb_000875_100 |
0.316413525 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 8 |
Hb_121031_010 |
0.3245867874 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 9 |
Hb_034050_010 |
0.3270202138 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 10 |
Hb_144101_010 |
0.3277627408 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 11 |
Hb_001014_200 |
0.335364363 |
- |
- |
- |
| 12 |
Hb_004162_300 |
0.3486286005 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 13 |
Hb_027014_010 |
0.3518226848 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 14 |
Hb_002352_040 |
0.3532288376 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 15 |
Hb_001544_020 |
0.3574834891 |
- |
- |
- |
| 16 |
Hb_002135_040 |
0.3584197244 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 17 |
Hb_002928_100 |
0.3590176957 |
- |
- |
- |
| 18 |
Hb_000931_030 |
0.3598681541 |
- |
- |
PREDICTED: uncharacterized protein LOC105130278 [Populus euphratica] |
| 19 |
Hb_029162_020 |
0.3604116606 |
- |
- |
- |
| 20 |
Hb_166548_010 |
0.3646303447 |
- |
- |
hypothetical protein JCGZ_09681 [Jatropha curcas] |