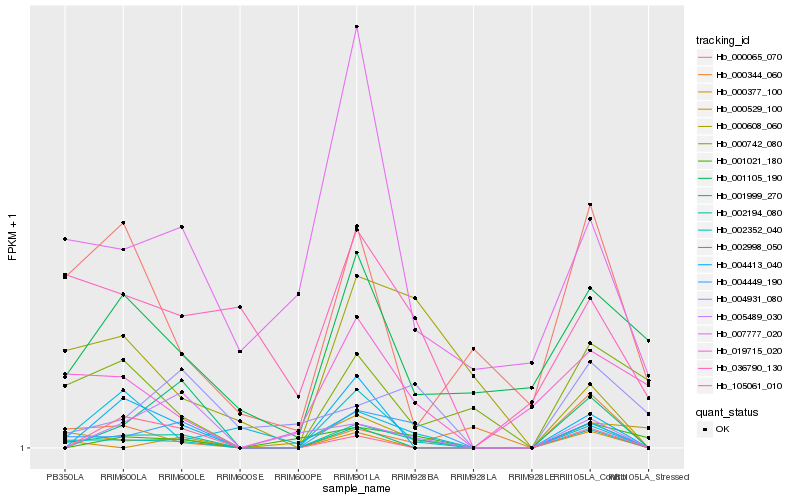
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002194_080 |
0.0 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004449_190 |
0.1696144577 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 3 |
Hb_002352_040 |
0.2812162308 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 4 |
Hb_004413_040 |
0.3352128937 |
- |
- |
- |
| 5 |
Hb_005489_030 |
0.3663227118 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 6 |
Hb_000377_100 |
0.3672204912 |
- |
- |
LINE-type retrotransposon LIb DNA, Insertion at the S11 site-like protein [Theobroma cacao] |
| 7 |
Hb_007777_020 |
0.3709284746 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 8 |
Hb_019715_020 |
0.371339503 |
- |
- |
- |
| 9 |
Hb_000065_070 |
0.3738233587 |
- |
- |
hypothetical protein CISIN_1g039077mg [Citrus sinensis] |
| 10 |
Hb_000529_100 |
0.3739723772 |
- |
- |
hypothetical protein POPTR_0009s05980g [Populus trichocarpa] |
| 11 |
Hb_001021_180 |
0.3754203747 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 12 |
Hb_105061_010 |
0.3765833864 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_000742_080 |
0.3773427836 |
- |
- |
- |
| 14 |
Hb_000344_060 |
0.3776991795 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 15 |
Hb_001105_190 |
0.3782079468 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 16 |
Hb_004931_080 |
0.3831248867 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Jatropha curcas] |
| 17 |
Hb_036790_130 |
0.384927432 |
- |
- |
- |
| 18 |
Hb_000608_060 |
0.385198145 |
- |
- |
- |
| 19 |
Hb_001999_270 |
0.3872764632 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 20 |
Hb_002998_050 |
0.3881707652 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Sesamum indicum] |