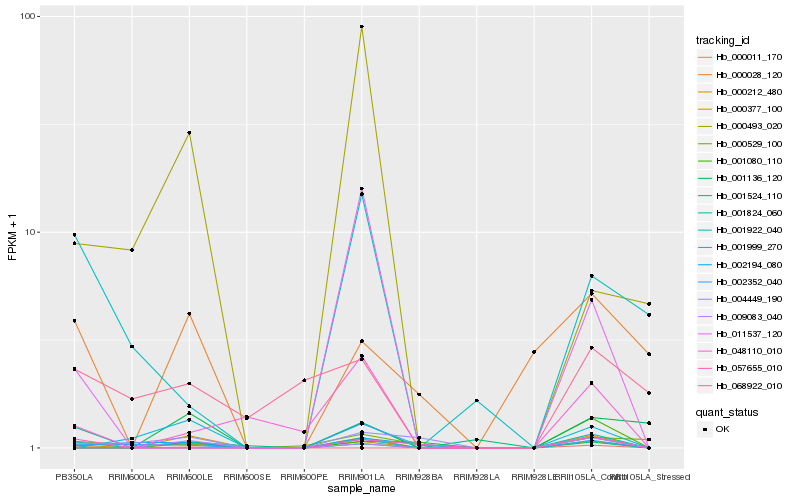
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000377_100 |
0.0 |
- |
- |
LINE-type retrotransposon LIb DNA, Insertion at the S11 site-like protein [Theobroma cacao] |
| 2 |
Hb_000212_480 |
0.2817523116 |
- |
- |
- |
| 3 |
Hb_009083_040 |
0.3117825572 |
- |
- |
hypothetical protein JCGZ_12563 [Jatropha curcas] |
| 4 |
Hb_002194_080 |
0.3672204912 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 5 |
Hb_048110_010 |
0.3741871185 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 6 |
Hb_004449_190 |
0.3890669095 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 7 |
Hb_002352_040 |
0.3957572353 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 8 |
Hb_001824_060 |
0.4040007552 |
- |
- |
- |
| 9 |
Hb_000028_120 |
0.4046459752 |
- |
- |
Os03g0221800 [Oryza sativa Japonica Group] |
| 10 |
Hb_001524_110 |
0.4279482123 |
- |
- |
PREDICTED: uncharacterized protein LOC104611877 isoform X2 [Nelumbo nucifera] |
| 11 |
Hb_057655_010 |
0.4311796644 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_001136_120 |
0.4336311942 |
- |
- |
- |
| 13 |
Hb_001999_270 |
0.4371281193 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 14 |
Hb_000011_170 |
0.4378288771 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 15 |
Hb_011537_120 |
0.4436272777 |
- |
- |
- |
| 16 |
Hb_000493_020 |
0.4439828242 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001922_040 |
0.44787895 |
- |
- |
reticulon family protein [Populus trichocarpa] |
| 18 |
Hb_068922_010 |
0.4480483336 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 19 |
Hb_001080_110 |
0.4487756294 |
- |
- |
- |
| 20 |
Hb_000529_100 |
0.4493130608 |
- |
- |
hypothetical protein POPTR_0009s05980g [Populus trichocarpa] |