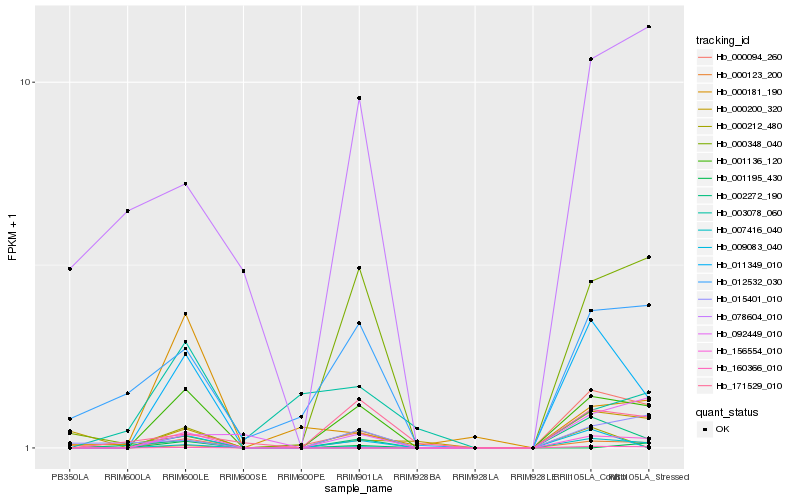
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001136_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_007416_040 |
0.2727606835 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_002272_190 |
0.3077472269 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_012532_030 |
0.3170487296 |
- |
- |
- |
| 5 |
Hb_171529_010 |
0.3287735712 |
- |
- |
- |
| 6 |
Hb_000212_480 |
0.3390449076 |
- |
- |
- |
| 7 |
Hb_156554_010 |
0.3411436102 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 8 |
Hb_015401_010 |
0.3479123251 |
- |
- |
hypothetical protein VITISV_001548 [Vitis vinifera] |
| 9 |
Hb_000094_260 |
0.352610211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009083_040 |
0.3602063539 |
- |
- |
hypothetical protein JCGZ_12563 [Jatropha curcas] |
| 11 |
Hb_092449_010 |
0.3652840941 |
- |
- |
- |
| 12 |
Hb_078604_010 |
0.3689708823 |
- |
- |
- |
| 13 |
Hb_011349_010 |
0.3733173274 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 14 |
Hb_000200_320 |
0.387083768 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_09311 [Jatropha curcas] |
| 15 |
Hb_000181_190 |
0.3907985079 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 16 |
Hb_000348_040 |
0.3975921599 |
- |
- |
- |
| 17 |
Hb_001195_430 |
0.3999163852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000123_200 |
0.4005921242 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 19 |
Hb_003078_060 |
0.4010440218 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 20 |
Hb_160366_010 |
0.4049473021 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |