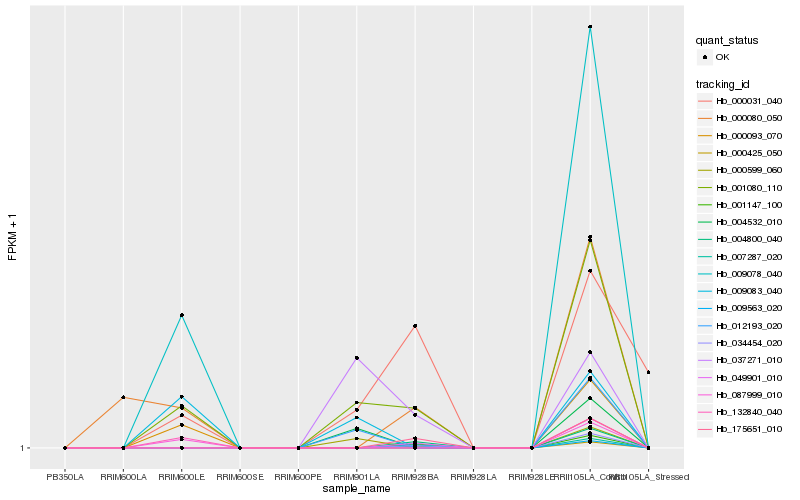
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_009083_040 |
0.3219887798 |
- |
- |
hypothetical protein JCGZ_12563 [Jatropha curcas] |
| 3 |
Hb_012193_020 |
0.3523196778 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 4 |
Hb_009078_040 |
0.3613371713 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 5 |
Hb_000080_050 |
0.361860661 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38150-like [Vitis vinifera] |
| 6 |
Hb_049901_010 |
0.3637497278 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 7 |
Hb_034454_020 |
0.3637648744 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_037271_010 |
0.3649333217 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 9 |
Hb_000031_040 |
0.3674483766 |
- |
- |
centromere protein O [Medicago truncatula] |
| 10 |
Hb_004532_010 |
0.3675127306 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 11 |
Hb_087999_010 |
0.3701408808 |
- |
- |
PREDICTED: uncharacterized protein LOC105640118 [Jatropha curcas] |
| 12 |
Hb_000093_070 |
0.3703078539 |
transcription factor |
TF Family: C2C2-YABBY |
Protein CRABS CLAW, putative [Ricinus communis] |
| 13 |
Hb_132840_040 |
0.372656528 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase, putative [Ricinus communis] |
| 14 |
Hb_000425_050 |
0.3757431837 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 15 |
Hb_009563_020 |
0.3757716681 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_007287_020 |
0.375825813 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 17 |
Hb_001147_100 |
0.3758986586 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 18 |
Hb_000599_060 |
0.3760240166 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 19 |
Hb_004800_040 |
0.3760705189 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 20 |
Hb_175651_010 |
0.3763228943 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |