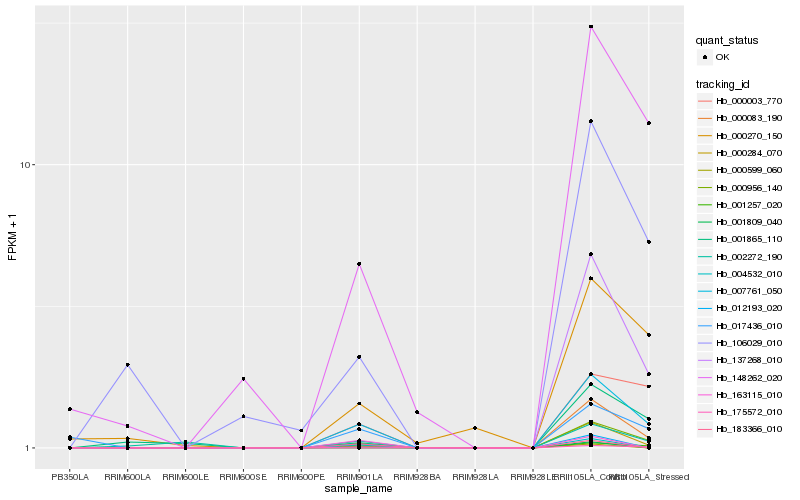
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 2 |
Hb_007761_050 |
0.1403734114 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 3 |
Hb_001257_020 |
0.2010480055 |
- |
- |
- |
| 4 |
Hb_012193_020 |
0.2106662193 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 5 |
Hb_004532_010 |
0.2449941938 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 6 |
Hb_000599_060 |
0.2612396692 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 7 |
Hb_148262_020 |
0.2678335663 |
- |
- |
- |
| 8 |
Hb_000083_190 |
0.2712964515 |
- |
- |
low affinity nitrate transporter, partial [Triticum aestivum] |
| 9 |
Hb_001809_040 |
0.2720627857 |
- |
- |
PREDICTED: uncharacterized protein LOC105638660 [Jatropha curcas] |
| 10 |
Hb_106029_010 |
0.2744450963 |
- |
- |
Transcription factor ICE1, putative [Ricinus communis] |
| 11 |
Hb_183366_010 |
0.28095465 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 12 |
Hb_137268_010 |
0.2813088611 |
- |
- |
- |
| 13 |
Hb_000956_140 |
0.2945727056 |
- |
- |
- |
| 14 |
Hb_175572_010 |
0.2975839785 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_163115_010 |
0.3104038264 |
- |
- |
- |
| 16 |
Hb_001865_110 |
0.3132531108 |
- |
- |
- |
| 17 |
Hb_000270_150 |
0.3170202957 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_017436_010 |
0.3180204718 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_002272_190 |
0.3252839979 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_000003_770 |
0.3256780355 |
- |
- |
- |