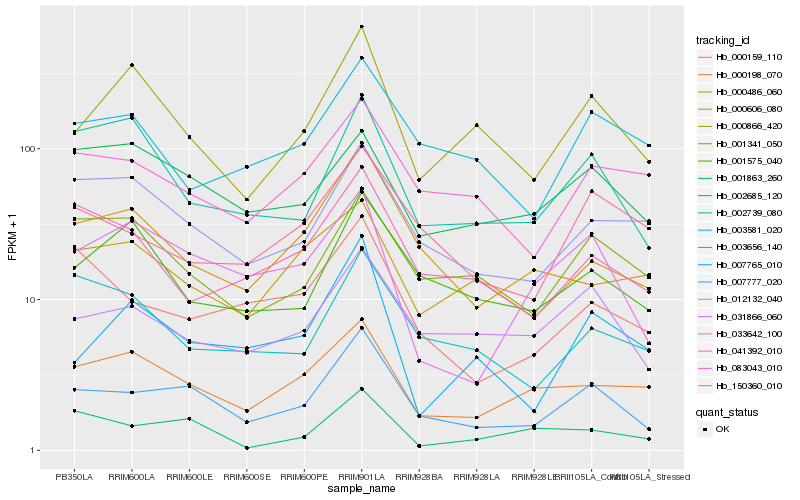
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007777_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 2 |
Hb_031866_060 |
0.1553511753 |
- |
- |
PREDICTED: uncharacterized protein LOC105642381 isoform X2 [Jatropha curcas] |
| 3 |
Hb_033642_100 |
0.1711895872 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Prunus mume] |
| 4 |
Hb_000866_420 |
0.1732646983 |
- |
- |
Remorin, putative [Ricinus communis] |
| 5 |
Hb_041392_010 |
0.1734823224 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 6 |
Hb_012132_040 |
0.1744192722 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 7 |
Hb_000159_110 |
0.1758627723 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 8 |
Hb_000486_060 |
0.1769753522 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 9 |
Hb_150360_010 |
0.1773470272 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 10 |
Hb_000198_070 |
0.1783314241 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 11 |
Hb_002739_080 |
0.1817445743 |
- |
- |
cystathionine beta-lyase, putative [Ricinus communis] |
| 12 |
Hb_083043_010 |
0.1826159222 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 13 |
Hb_001341_050 |
0.1837484252 |
- |
- |
ATP synthase D chain, mitochondrial [Theobroma cacao] |
| 14 |
Hb_000606_080 |
0.1846197162 |
- |
- |
PREDICTED: RNA-binding protein Nova-2-like [Malus domestica] |
| 15 |
Hb_001575_040 |
0.1855619592 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003581_020 |
0.188001414 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003656_140 |
0.1889685233 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007765_010 |
0.1891029332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002685_120 |
0.1896967592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001863_260 |
0.1897584128 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |