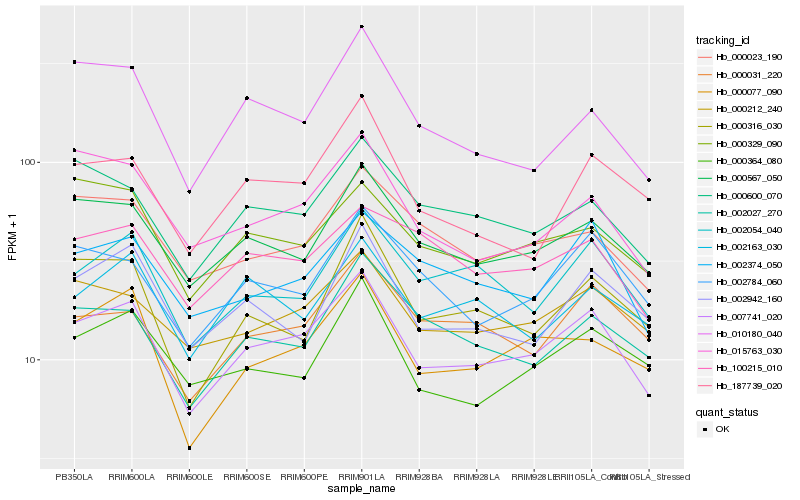
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007741_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 2 |
Hb_000567_050 |
0.0813016083 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 3 |
Hb_000077_090 |
0.0865728094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000212_240 |
0.0878159331 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_002054_040 |
0.0895020146 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 6 |
Hb_002374_050 |
0.0907732454 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 7 |
Hb_002784_060 |
0.0938187346 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000600_070 |
0.0967542808 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_000023_190 |
0.0973305199 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 10 |
Hb_000031_220 |
0.0981749944 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 11 |
Hb_000364_080 |
0.0985113835 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002027_270 |
0.0992317748 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 13 |
Hb_015763_030 |
0.0997509411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002942_160 |
0.1001453959 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 15 |
Hb_000316_030 |
0.1022912403 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000329_090 |
0.1023623641 |
- |
- |
arginase, putative [Ricinus communis] |
| 17 |
Hb_187739_020 |
0.1031751074 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 18 |
Hb_100215_010 |
0.1034153169 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 19 |
Hb_002163_030 |
0.1038379664 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 59 kDa regulatory subunit B' gamma isoform [Jatropha curcas] |
| 20 |
Hb_010180_040 |
0.1048402878 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |