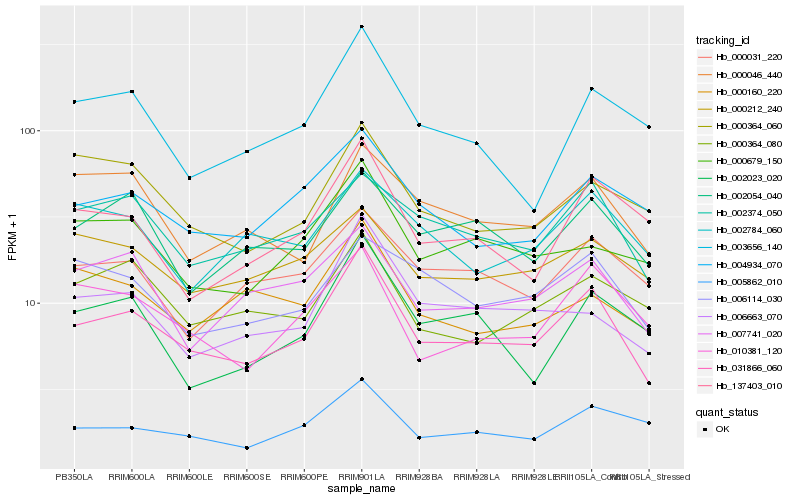
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031866_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642381 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002374_050 |
0.1182831721 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 3 |
Hb_004934_070 |
0.1195865774 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 4 |
Hb_006663_070 |
0.1200900607 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 5 |
Hb_005862_010 |
0.121215831 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 6 |
Hb_010381_120 |
0.1215763669 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 7 |
Hb_007741_020 |
0.1220946086 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 8 |
Hb_137403_010 |
0.1261638402 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 9 |
Hb_000679_150 |
0.1272169708 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_003656_140 |
0.1281935813 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002054_040 |
0.1282868179 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 12 |
Hb_000031_220 |
0.1295572167 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 13 |
Hb_002023_020 |
0.1298351244 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 14 |
Hb_000364_080 |
0.1306210991 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000046_440 |
0.1322266781 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 16 |
Hb_006114_030 |
0.1337435123 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 17 |
Hb_000364_060 |
0.1341650747 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 18 |
Hb_002784_060 |
0.1357460515 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000160_220 |
0.1364484147 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000212_240 |
0.1370474263 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |