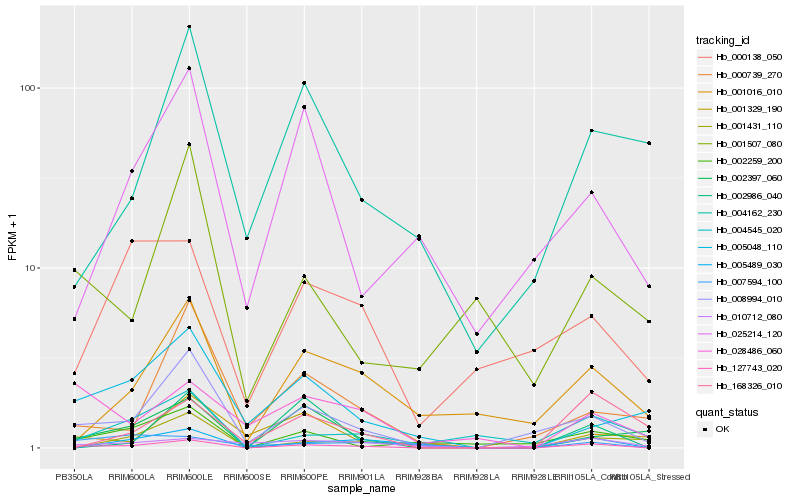
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127743_020 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 2 |
Hb_008994_010 |
0.2378582454 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_005489_030 |
0.3002027532 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 4 |
Hb_002397_060 |
0.3105466646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004545_020 |
0.3173013596 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 6 |
Hb_001016_010 |
0.3290742733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002259_200 |
0.3323378574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005048_110 |
0.3328817844 |
- |
- |
- |
| 9 |
Hb_002986_040 |
0.3347423068 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 10 |
Hb_028486_060 |
0.3374623981 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 11 |
Hb_000739_270 |
0.3408521886 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 12 |
Hb_007594_100 |
0.3425130834 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 13 |
Hb_001507_080 |
0.348312516 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 14 |
Hb_025214_120 |
0.3502417431 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 15 |
Hb_168326_010 |
0.350614688 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 16 |
Hb_000138_050 |
0.352480892 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper protein HAT14, putative [Ricinus communis] |
| 17 |
Hb_010712_080 |
0.354372181 |
- |
- |
hypothetical protein AALP_AA5G019400 [Arabis alpina] |
| 18 |
Hb_004162_230 |
0.3562865333 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 19 |
Hb_001329_190 |
0.3565206142 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_001431_110 |
0.3589939537 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |