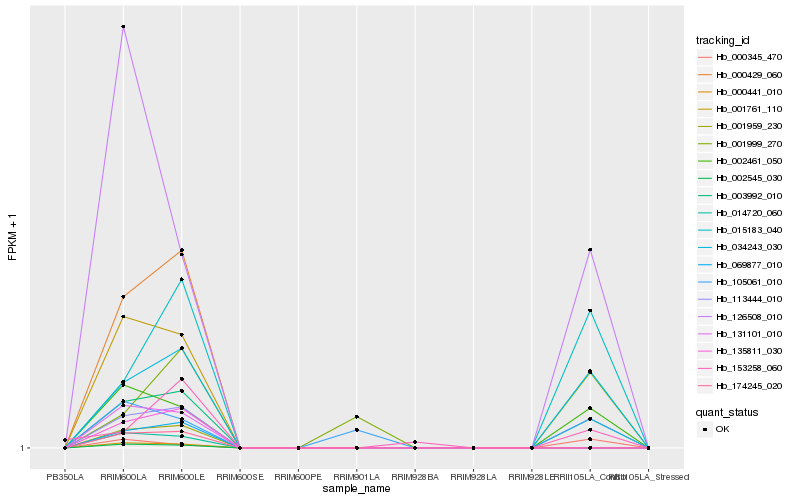
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_034243_030 |
0.1070118228 |
- |
- |
B3 domain-containing protein [Medicago truncatula] |
| 3 |
Hb_002461_050 |
0.1389372493 |
- |
- |
- |
| 4 |
Hb_015183_040 |
0.2050512679 |
- |
- |
- |
| 5 |
Hb_126508_010 |
0.2507769771 |
- |
- |
- |
| 6 |
Hb_000345_470 |
0.2882536063 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 7 |
Hb_105061_010 |
0.3093852519 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 8 |
Hb_001999_270 |
0.3236647167 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 9 |
Hb_153258_060 |
0.3289501133 |
- |
- |
- |
| 10 |
Hb_001959_230 |
0.3404217982 |
- |
- |
PREDICTED: uncharacterized protein LOC105638436 [Jatropha curcas] |
| 11 |
Hb_113444_010 |
0.3404875333 |
- |
- |
- |
| 12 |
Hb_174245_020 |
0.3410677868 |
- |
- |
- |
| 13 |
Hb_000429_060 |
0.3412563789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_069877_010 |
0.3427852752 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 15 |
Hb_135811_030 |
0.3433240469 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_001761_110 |
0.3495617269 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 17 |
Hb_131101_010 |
0.350516122 |
- |
- |
- |
| 18 |
Hb_014720_060 |
0.3542050344 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 19 |
Hb_000441_010 |
0.3543930151 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_002545_030 |
0.3557033042 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |