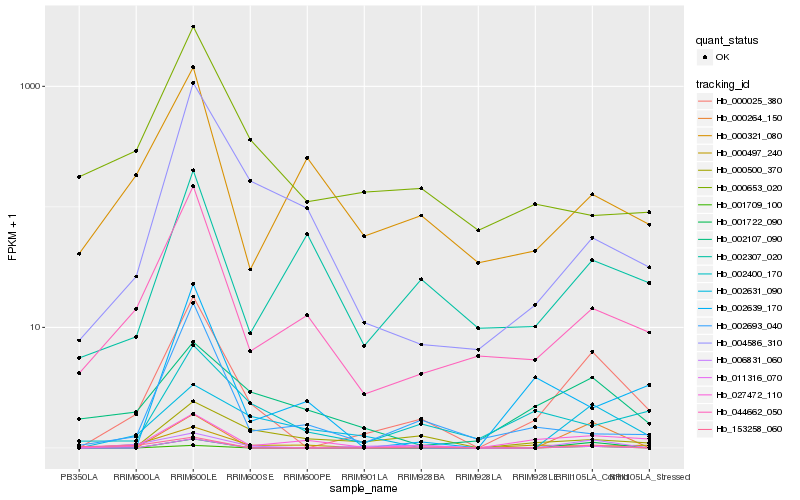
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_380 |
0.0 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 2 |
Hb_006831_060 |
0.2674662586 |
- |
- |
hypothetical protein JCGZ_16419 [Jatropha curcas] |
| 3 |
Hb_044662_050 |
0.2819647168 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 4 |
Hb_027472_110 |
0.2840848543 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 5 |
Hb_004586_310 |
0.3217677247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002400_170 |
0.3241896352 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 7 |
Hb_002693_040 |
0.3259136586 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 8 |
Hb_002631_090 |
0.3276051231 |
- |
- |
- |
| 9 |
Hb_011316_070 |
0.3292466935 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000497_240 |
0.3306511955 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 11 |
Hb_002107_090 |
0.3310663176 |
transcription factor |
TF Family: bZIP |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 12 |
Hb_000653_020 |
0.33128595 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 13 |
Hb_001709_100 |
0.337010345 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 14 |
Hb_001722_090 |
0.3372989244 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 15 |
Hb_000500_370 |
0.3388776542 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000321_080 |
0.3432417852 |
- |
- |
histone H4 [Zea mays] |
| 17 |
Hb_002639_170 |
0.346426584 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 18 |
Hb_153258_060 |
0.3480140978 |
- |
- |
- |
| 19 |
Hb_000264_150 |
0.3486154154 |
- |
- |
PREDICTED: uncharacterized protein LOC103870759 [Brassica rapa] |
| 20 |
Hb_002307_020 |
0.3505422275 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |