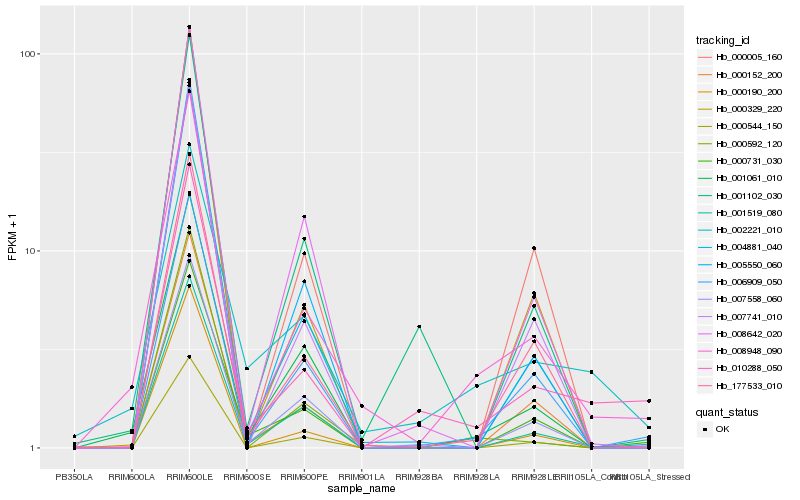
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_090 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001061_010 |
0.1396469179 |
- |
- |
PREDICTED: uncharacterized protein LOC100243595 [Vitis vinifera] |
| 3 |
Hb_006909_050 |
0.1450987354 |
transcription factor |
TF Family: OFP |
hypothetical protein POPTR_0008s01840g [Populus trichocarpa] |
| 4 |
Hb_177533_010 |
0.1520631449 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 5 |
Hb_001519_080 |
0.1548123849 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 6 |
Hb_004881_040 |
0.1583680324 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 7 |
Hb_000592_120 |
0.1625044422 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 8 |
Hb_010288_050 |
0.1664823974 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 9 |
Hb_000544_150 |
0.1668098534 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 10 |
Hb_008642_020 |
0.1669240107 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 11 |
Hb_007741_010 |
0.1689497834 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 12 |
Hb_002221_010 |
0.1702602277 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 13 |
Hb_000152_200 |
0.1704472522 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000731_030 |
0.1710541622 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_007558_060 |
0.1712758614 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 16 |
Hb_000005_160 |
0.1715657878 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 17 |
Hb_005550_060 |
0.171859737 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000190_200 |
0.173169514 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 19 |
Hb_000329_220 |
0.173334063 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 20 |
Hb_001102_030 |
0.1749259638 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |