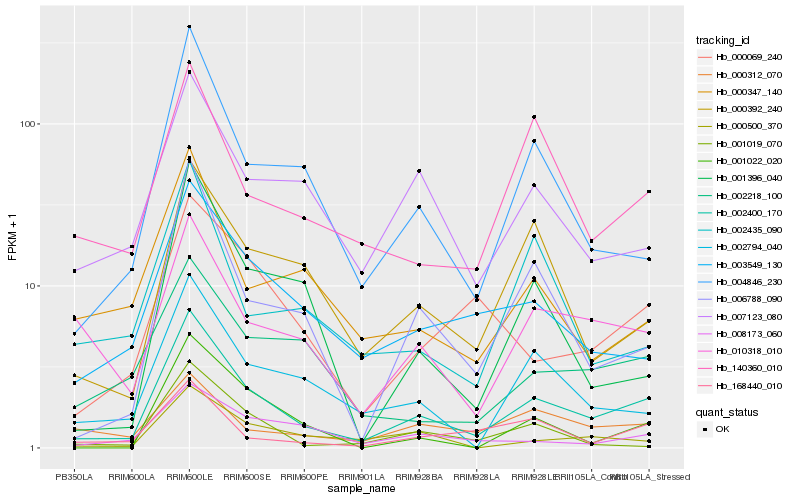
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_170 |
0.0 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 2 |
Hb_006788_090 |
0.1794662203 |
- |
- |
- |
| 3 |
Hb_004846_230 |
0.1836106479 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_002794_040 |
0.1954128366 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 5 |
Hb_010318_010 |
0.1983367321 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003549_130 |
0.1983867071 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 7 |
Hb_000392_240 |
0.200976595 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_007123_080 |
0.2040298425 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 9 |
Hb_140360_010 |
0.2067292363 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_168440_010 |
0.212042553 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 11 |
Hb_002218_100 |
0.2127056861 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 12 |
Hb_000069_240 |
0.2148683354 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 13 |
Hb_000500_370 |
0.2151974033 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_000347_140 |
0.2168558753 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_008173_060 |
0.2213628213 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_001396_040 |
0.2224193024 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 17 |
Hb_001022_020 |
0.222998559 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 18 |
Hb_000312_070 |
0.2233022301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002435_090 |
0.2256979871 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001019_070 |
0.2276355813 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |