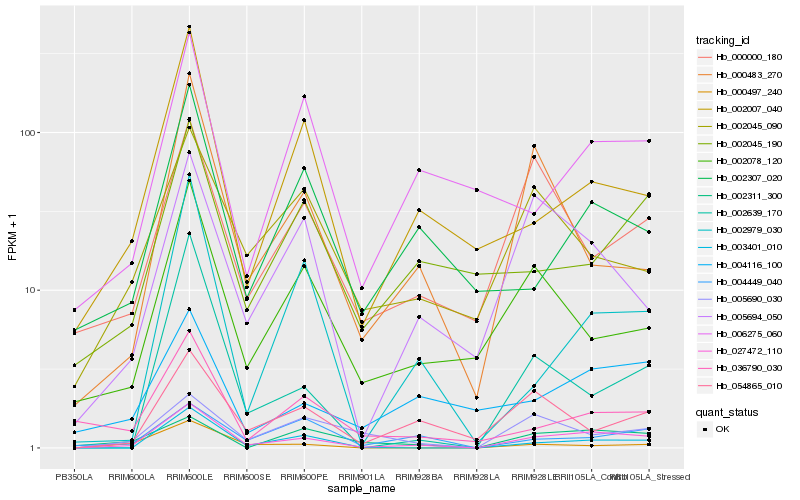
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027472_110 |
0.0 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 2 |
Hb_003401_010 |
0.1958112035 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 3 |
Hb_005694_050 |
0.2307901456 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_054865_010 |
0.2381764102 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 5 |
Hb_004116_100 |
0.2389222514 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 6 |
Hb_002078_120 |
0.2395351926 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 7 |
Hb_002007_040 |
0.2395427564 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_002307_020 |
0.2410976301 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 9 |
Hb_002979_030 |
0.2442179269 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_036790_030 |
0.2455804799 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 11 |
Hb_002639_170 |
0.2460665615 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 12 |
Hb_000483_270 |
0.2488988386 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 13 |
Hb_000000_180 |
0.2490430627 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_006275_060 |
0.2526524783 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 15 |
Hb_002045_190 |
0.253938348 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 16 |
Hb_000497_240 |
0.2577606446 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 17 |
Hb_002045_090 |
0.2579482497 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 18 |
Hb_005690_030 |
0.2591751477 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 19 |
Hb_004449_040 |
0.2593247313 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 20 |
Hb_002311_300 |
0.2604432298 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |