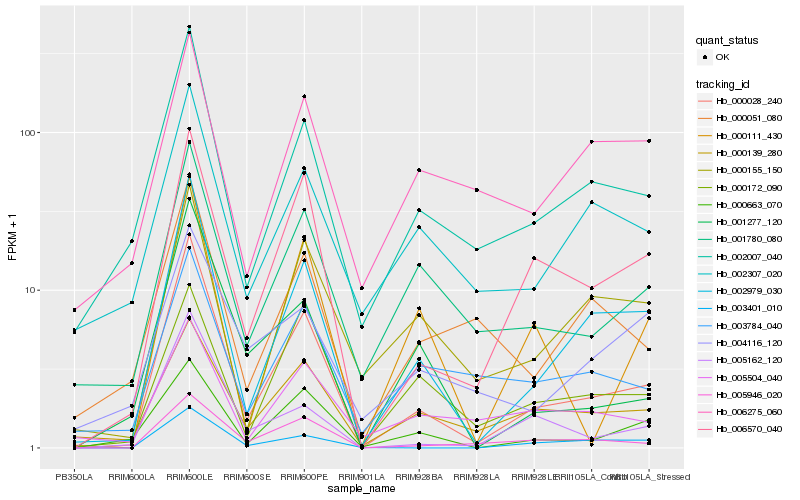
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002979_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000028_240 |
0.1075797958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002007_040 |
0.1636568355 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000155_150 |
0.1727345826 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 5 |
Hb_003401_010 |
0.1778948484 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 6 |
Hb_006570_040 |
0.1881434033 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 7 |
Hb_000663_070 |
0.1911640994 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 8 |
Hb_002307_020 |
0.1959149632 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 9 |
Hb_000051_080 |
0.2085420443 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 10 |
Hb_001780_080 |
0.2145932664 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 11 |
Hb_000139_280 |
0.2149941276 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006275_060 |
0.2158809201 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 13 |
Hb_005946_020 |
0.2182628995 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |
| 14 |
Hb_000172_090 |
0.2274583888 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 15 |
Hb_003784_040 |
0.228497869 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 16 |
Hb_001277_120 |
0.2285396298 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 17 |
Hb_005162_120 |
0.2303049838 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 18 |
Hb_004116_120 |
0.2309536454 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 19 |
Hb_000111_430 |
0.2345427951 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 20 |
Hb_005504_040 |
0.2359866259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |