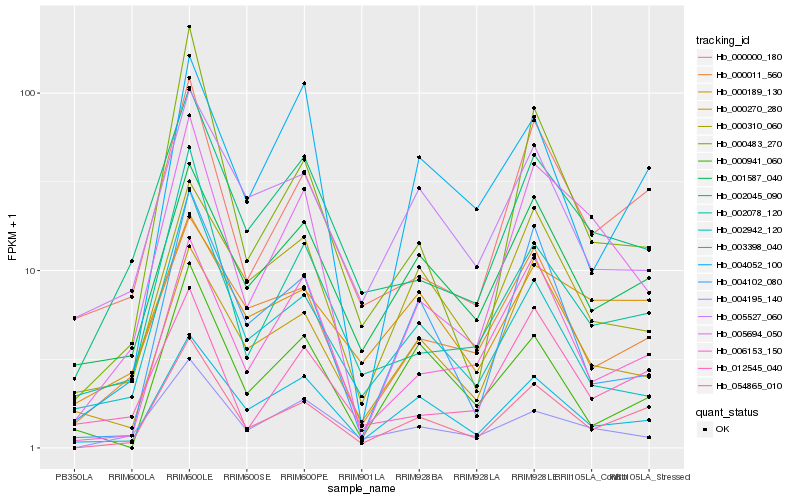
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 2 |
Hb_000000_180 |
0.1432196527 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_003398_040 |
0.1505011173 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001587_040 |
0.1612297061 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002078_120 |
0.1619298676 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 6 |
Hb_000011_560 |
0.1682071647 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002045_090 |
0.1774901253 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 8 |
Hb_000483_270 |
0.178427029 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 9 |
Hb_004102_080 |
0.178781222 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 10 |
Hb_006153_150 |
0.1813262983 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 11 |
Hb_000310_060 |
0.1830206961 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 12 |
Hb_000941_060 |
0.183401108 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 13 |
Hb_004195_140 |
0.1844721981 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 14 |
Hb_004052_100 |
0.1850820594 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 15 |
Hb_000270_280 |
0.1855871178 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 16 |
Hb_002942_120 |
0.1872881645 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 17 |
Hb_012545_040 |
0.1888207918 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_005694_050 |
0.1895676146 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_000189_130 |
0.1915965168 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005527_060 |
0.191677088 |
- |
- |
malic enzyme, putative [Ricinus communis] |