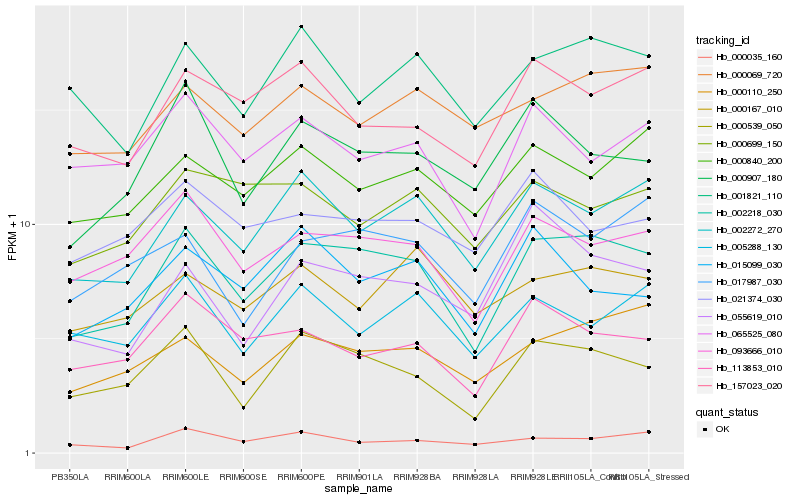
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002218_030 |
0.0 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 2 |
Hb_000539_050 |
0.0886590335 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 3 |
Hb_093666_010 |
0.0988429668 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 4 |
Hb_002272_270 |
0.10356731 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 5 |
Hb_000110_250 |
0.1129733892 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |
| 6 |
Hb_000907_180 |
0.1163709613 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 7 |
Hb_113853_010 |
0.1180274716 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 8 |
Hb_157023_020 |
0.1191523957 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001821_110 |
0.1201342365 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000840_200 |
0.1237048837 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000069_720 |
0.1248141704 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 12 |
Hb_065525_080 |
0.1249089187 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 13 |
Hb_017987_030 |
0.1256724718 |
- |
- |
PREDICTED: uncharacterized protein LOC105630796 [Jatropha curcas] |
| 14 |
Hb_000035_160 |
0.1281898219 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 15 |
Hb_015099_030 |
0.1288986731 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 16 |
Hb_000699_150 |
0.1311386299 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 17 |
Hb_055619_010 |
0.1317237294 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000167_010 |
0.1319377426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005288_130 |
0.1329507985 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_021374_030 |
0.1342316174 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |