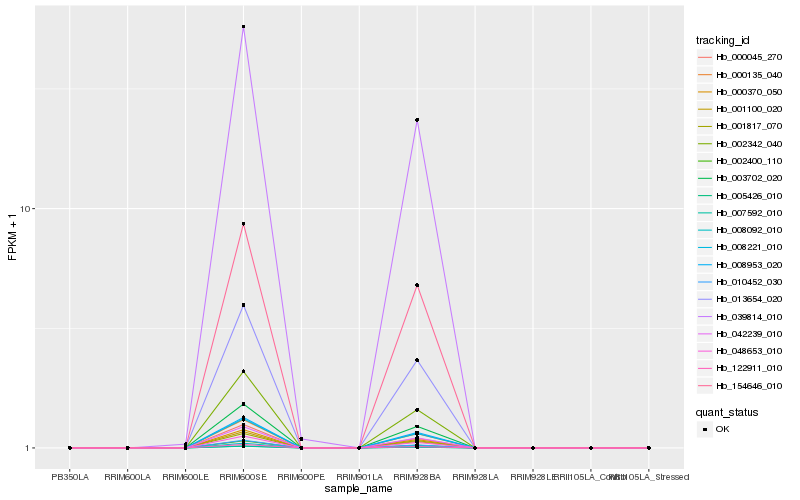
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005426_010 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_007592_010 |
0.000154192 |
- |
- |
PREDICTED: uncharacterized protein LOC105118874 [Populus euphratica] |
| 3 |
Hb_008092_010 |
0.0023008313 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 4 |
Hb_003702_020 |
0.002744712 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 5 |
Hb_000370_050 |
0.0027802457 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_042239_010 |
0.0070585147 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 7 |
Hb_048653_010 |
0.0083211781 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 8 |
Hb_013654_020 |
0.008463453 |
- |
- |
putative prolin-rich extensin-like receptor protein kinase family protein [Zea mays] |
| 9 |
Hb_002342_040 |
0.0103864321 |
- |
- |
FERONIA, partial [Arabis alpina] |
| 10 |
Hb_008221_010 |
0.0120998888 |
- |
- |
hypothetical protein JCGZ_10961 [Jatropha curcas] |
| 11 |
Hb_008953_020 |
0.0170207288 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 12 |
Hb_000135_040 |
0.017212741 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At5g56810 [Jatropha curcas] |
| 13 |
Hb_122911_010 |
0.017589414 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000045_270 |
0.0232751244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001817_070 |
0.0248450318 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_010452_030 |
0.025966641 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 17 |
Hb_001100_020 |
0.0272841607 |
- |
- |
- |
| 18 |
Hb_154646_010 |
0.0277455833 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_002400_110 |
0.0324631027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_039814_010 |
0.0325704662 |
- |
- |
hypothetical protein JCGZ_05023 [Jatropha curcas] |