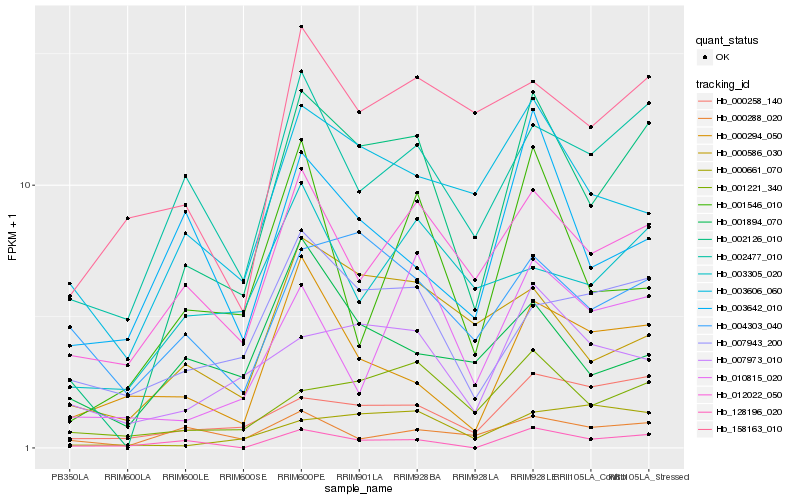
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002126_010 |
0.0 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 2 |
Hb_002477_010 |
0.1862436851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001221_340 |
0.18627683 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 4 |
Hb_012022_050 |
0.1872323368 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 5 |
Hb_000258_140 |
0.1940716032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007943_200 |
0.1970311451 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_007973_010 |
0.2053107104 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000586_030 |
0.2054167691 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 9 |
Hb_003606_060 |
0.2057875263 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 10 |
Hb_158163_010 |
0.2174615542 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_004303_040 |
0.2189178381 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003305_020 |
0.2217288128 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 13 |
Hb_000661_070 |
0.2218312967 |
- |
- |
- |
| 14 |
Hb_010815_020 |
0.2268616996 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_128196_020 |
0.2271682352 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Nelumbo nucifera] |
| 16 |
Hb_001894_070 |
0.2324226867 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000294_050 |
0.232516489 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 18 |
Hb_000288_020 |
0.2334553129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003642_010 |
0.2342706677 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_001546_010 |
0.235301359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |