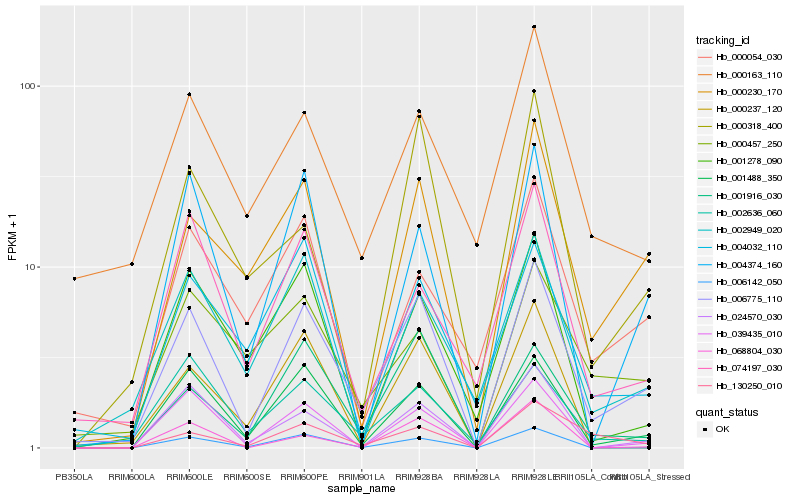
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006775_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 2 |
Hb_001488_350 |
0.1496329553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000230_170 |
0.171896912 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 4 |
Hb_024570_030 |
0.1820618752 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_039435_010 |
0.1854613937 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 6 |
Hb_001916_030 |
0.1868474351 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 7 |
Hb_004374_160 |
0.1887548275 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 8 |
Hb_002949_020 |
0.1919718994 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 9 |
Hb_002636_060 |
0.1935171591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006142_050 |
0.1974754339 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 11 |
Hb_130250_010 |
0.1978401162 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 12 |
Hb_000163_110 |
0.2016585425 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 13 |
Hb_074197_030 |
0.2018731019 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000237_120 |
0.204627138 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 15 |
Hb_000457_250 |
0.207660009 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 16 |
Hb_004032_110 |
0.2080083153 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001278_090 |
0.2081983089 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 18 |
Hb_000318_400 |
0.2091723049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000054_030 |
0.2104380848 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 20 |
Hb_068804_030 |
0.2107853946 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |