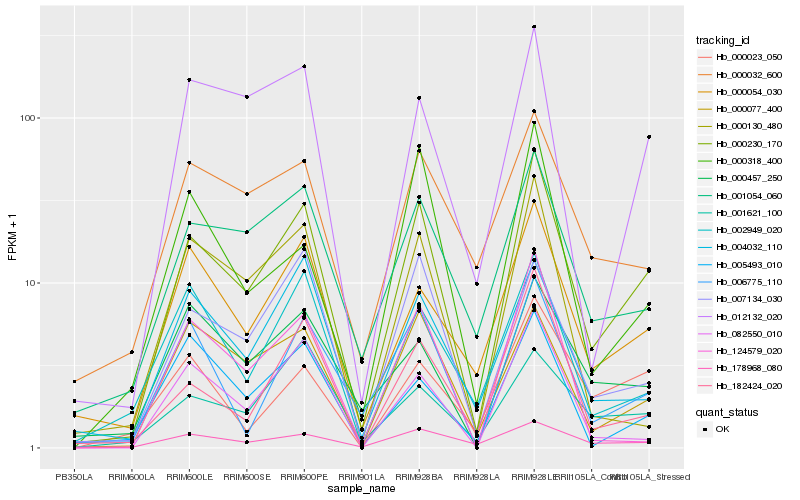
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_170 |
0.0 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 2 |
Hb_001054_060 |
0.1640415401 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 3 |
Hb_182424_020 |
0.1645566321 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 4 |
Hb_000054_030 |
0.1672382528 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 5 |
Hb_178968_080 |
0.1698828892 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012132_020 |
0.1702091321 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 7 |
Hb_006775_110 |
0.171896912 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 8 |
Hb_000077_400 |
0.1749178833 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_082550_010 |
0.1783406925 |
- |
- |
hypothetical protein CISIN_1g0081081mg, partial [Citrus sinensis] |
| 10 |
Hb_007134_030 |
0.1827837718 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 11 |
Hb_124579_020 |
0.1871767349 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 12 |
Hb_002949_020 |
0.1873926872 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 13 |
Hb_000032_600 |
0.18933429 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 14 |
Hb_000130_480 |
0.1895915356 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 15 |
Hb_000023_050 |
0.1913555899 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 16 |
Hb_004032_110 |
0.1932393287 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000318_400 |
0.1982358393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000457_250 |
0.1984932537 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 19 |
Hb_005493_010 |
0.1992692488 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_001621_100 |
0.2004734031 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |