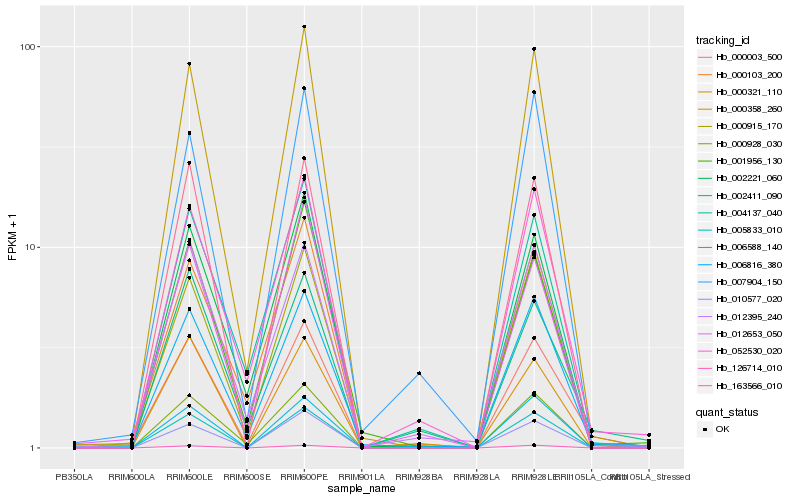
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_500 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 2 |
Hb_006816_380 |
0.0984463864 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 3 |
Hb_000358_260 |
0.1057692542 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 4 |
Hb_052530_020 |
0.1135415119 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 5 |
Hb_000928_030 |
0.1135506548 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 6 |
Hb_012395_240 |
0.1143957154 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005833_010 |
0.1154765549 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002411_090 |
0.1164410589 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 9 |
Hb_163566_010 |
0.1186266922 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 10 |
Hb_000915_170 |
0.1202890496 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_000103_200 |
0.1237691604 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_010577_020 |
0.1257108033 |
- |
- |
- |
| 13 |
Hb_004137_040 |
0.1267368769 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 14 |
Hb_000321_110 |
0.1271293805 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 15 |
Hb_126714_010 |
0.1271564306 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 16 |
Hb_006588_140 |
0.1295063895 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_007904_150 |
0.1301216559 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 18 |
Hb_001956_130 |
0.1318889549 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_012653_050 |
0.1320828363 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 20 |
Hb_002221_060 |
0.1321605723 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |