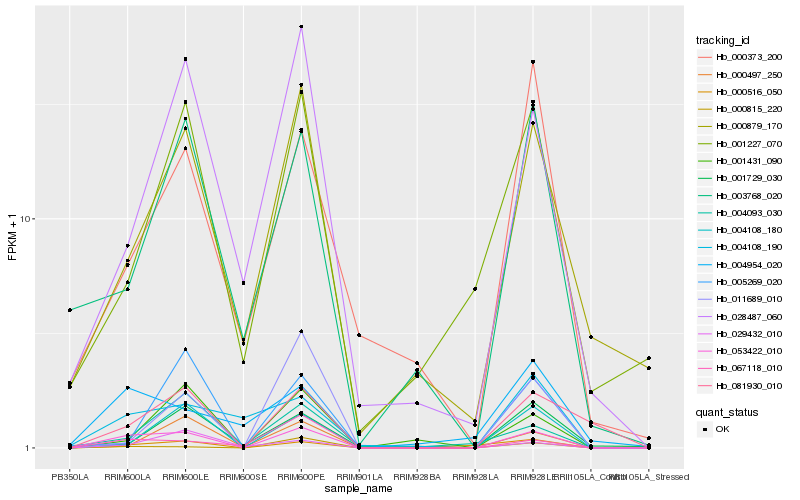
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053422_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 2 |
Hb_000879_170 |
0.2583100497 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_081930_010 |
0.2583722053 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 4 |
Hb_004108_180 |
0.2635365031 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 5 |
Hb_028487_060 |
0.2684160053 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 6 |
Hb_003768_020 |
0.2833587814 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 7 |
Hb_001431_090 |
0.2866083421 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 47, partial [Camelina sativa] |
| 8 |
Hb_001227_070 |
0.2896900801 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 9 |
Hb_067118_010 |
0.2898587665 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 10 |
Hb_004093_030 |
0.2937117475 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 11 |
Hb_000815_220 |
0.2948375127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004108_190 |
0.2949038575 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 13 |
Hb_011689_010 |
0.2996043007 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 14 |
Hb_029432_010 |
0.2996979131 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000497_250 |
0.3024718853 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 16 |
Hb_000373_200 |
0.3032222538 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 17 |
Hb_004954_020 |
0.3035322145 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005269_020 |
0.3051058302 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 19 |
Hb_000516_050 |
0.3073766855 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 20 |
Hb_001729_030 |
0.3122229587 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |