| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
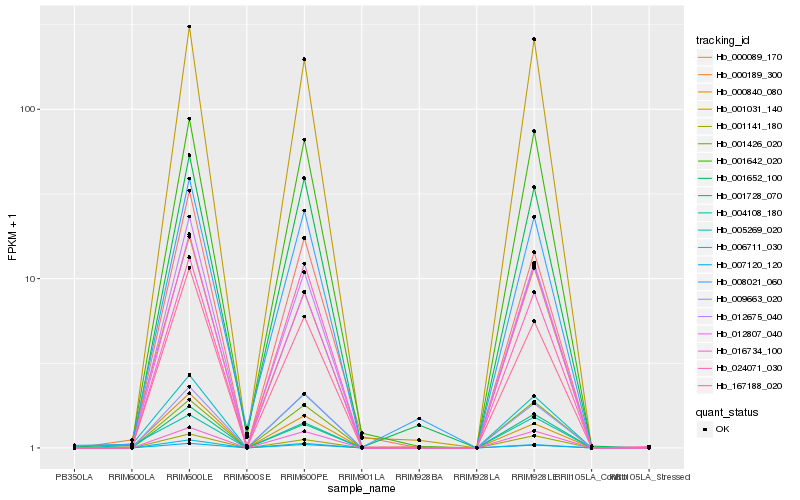
Hb_005269_020 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 2 |
Hb_004108_180 |
0.0981571046 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 3 |
Hb_007120_120 |
0.1003086888 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000189_300 |
0.1049896038 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000840_080 |
0.1069381356 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 6 |
Hb_008021_060 |
0.1074641078 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_009663_020 |
0.1075916044 |
- |
- |
- |
| 8 |
Hb_016734_100 |
0.1089406161 |
- |
- |
- |
| 9 |
Hb_001652_100 |
0.1096686933 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 10 |
Hb_001642_020 |
0.111538414 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 11 |
Hb_012675_040 |
0.1154663681 |
- |
- |
PREDICTED: uncharacterized protein LOC105647773 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001031_140 |
0.116050895 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 13 |
Hb_006711_030 |
0.1164127108 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 14 |
Hb_167188_020 |
0.1167168039 |
- |
- |
PREDICTED: uncharacterized protein At1g05835 [Jatropha curcas] |
| 15 |
Hb_001728_070 |
0.1211749499 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 16 |
Hb_024071_030 |
0.121618995 |
- |
- |
hypothetical protein CICLE_v10025479mg [Citrus clementina] |
| 17 |
Hb_000089_170 |
0.1218986688 |
- |
- |
PREDICTED: uncharacterized protein LOC105134595 [Populus euphratica] |
| 18 |
Hb_012807_040 |
0.1226714582 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 19 |
Hb_001426_020 |
0.1244509903 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 20 |
Hb_001141_180 |
0.124592765 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |