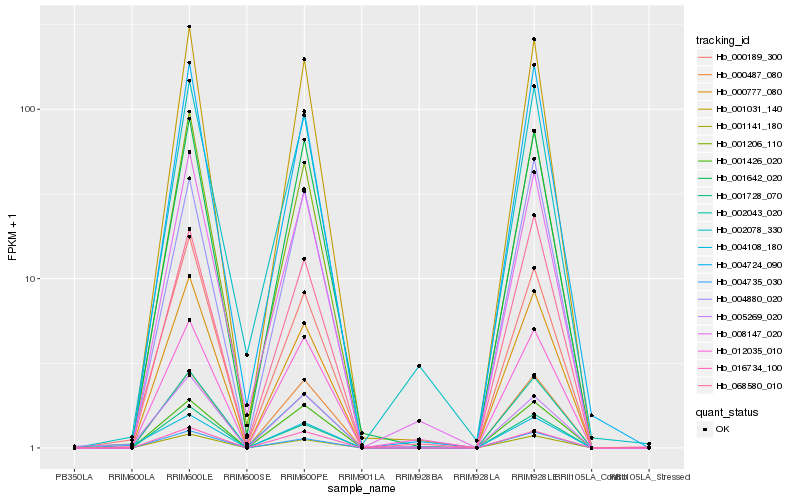
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004108_180 |
0.0 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 2 |
Hb_005269_020 |
0.0981571046 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 3 |
Hb_001642_020 |
0.1109996359 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 4 |
Hb_001031_140 |
0.1116103624 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 5 |
Hb_001426_020 |
0.1127778742 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 6 |
Hb_016734_100 |
0.1138796882 |
- |
- |
- |
| 7 |
Hb_001141_180 |
0.114718892 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 8 |
Hb_000487_080 |
0.11474778 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 9 |
Hb_000189_300 |
0.122550433 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001728_070 |
0.1239867786 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 11 |
Hb_004735_030 |
0.1244050838 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 12 |
Hb_002043_020 |
0.1257092077 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 13 |
Hb_000777_080 |
0.1277715551 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 14 |
Hb_001206_110 |
0.1281752832 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 15 |
Hb_004880_020 |
0.1284872494 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Jatropha curcas] |
| 16 |
Hb_068580_010 |
0.129590892 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 17 |
Hb_012035_010 |
0.1304845206 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 18 |
Hb_008147_020 |
0.1315087148 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 19 |
Hb_002078_330 |
0.1329964151 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_004724_090 |
0.1332345197 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |