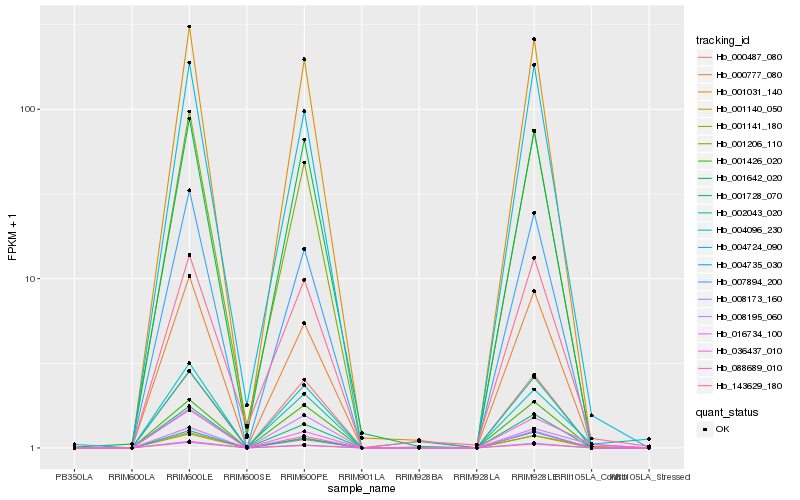
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_050 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1 [Jatropha curcas] |
| 2 |
Hb_008195_060 |
0.1349109547 |
transcription factor |
TF Family: NAC |
NAC transcription factor 018 [Jatropha curcas] |
| 3 |
Hb_001031_140 |
0.1389664142 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 4 |
Hb_143629_180 |
0.1402071274 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 5 |
Hb_001141_180 |
0.1403525508 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 6 |
Hb_001728_070 |
0.1421650808 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 7 |
Hb_036437_010 |
0.1423101958 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 8 |
Hb_016734_100 |
0.1436187545 |
- |
- |
- |
| 9 |
Hb_004724_090 |
0.1438599986 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 10 |
Hb_001642_020 |
0.1448251368 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 11 |
Hb_000777_080 |
0.1455724243 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 12 |
Hb_001206_110 |
0.1456744571 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 13 |
Hb_004735_030 |
0.1474177456 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 14 |
Hb_001426_020 |
0.1480586632 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 15 |
Hb_000487_080 |
0.1487877138 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 16 |
Hb_002043_020 |
0.1497662208 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 17 |
Hb_004096_230 |
0.1499054179 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 18 |
Hb_088689_010 |
0.1503823766 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 19 |
Hb_008173_160 |
0.1504244215 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 20 |
Hb_007894_200 |
0.150741181 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |