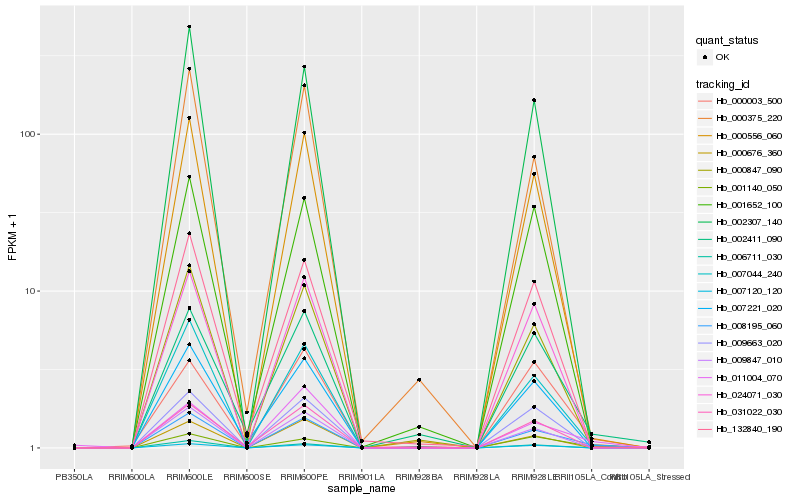
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008195_060 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 018 [Jatropha curcas] |
| 2 |
Hb_007044_240 |
0.1057715775 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000556_060 |
0.1160384508 |
- |
- |
hypothetical protein OsJ_28181 [Oryza sativa Japonica Group] |
| 4 |
Hb_009847_010 |
0.1236093157 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 5 |
Hb_024071_030 |
0.1319529614 |
- |
- |
hypothetical protein CICLE_v10025479mg [Citrus clementina] |
| 6 |
Hb_000847_090 |
0.13393608 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001140_050 |
0.1349109547 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1 [Jatropha curcas] |
| 8 |
Hb_009663_020 |
0.1364459431 |
- |
- |
- |
| 9 |
Hb_007120_120 |
0.1384706955 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_132840_190 |
0.1410770518 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_031022_030 |
0.141785654 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 12 |
Hb_006711_030 |
0.1428513265 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 13 |
Hb_002307_140 |
0.1437461104 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |
| 14 |
Hb_007221_020 |
0.1442166627 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_002411_090 |
0.1447576484 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 16 |
Hb_011004_070 |
0.1476912701 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 17 |
Hb_000676_360 |
0.1479566249 |
- |
- |
hypothetical protein POPTR_0007s13690g [Populus trichocarpa] |
| 18 |
Hb_000003_500 |
0.1485917266 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 19 |
Hb_000375_220 |
0.1492070633 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 20 |
Hb_001652_100 |
0.1495592755 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |