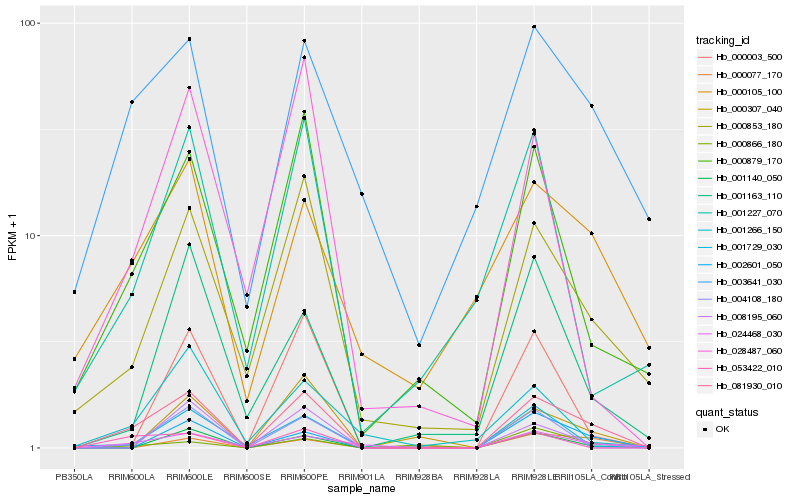
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081930_010 |
0.0 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 2 |
Hb_000853_180 |
0.2326159613 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 3 |
Hb_000879_170 |
0.2353648396 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_024468_030 |
0.245412891 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 5 |
Hb_001140_050 |
0.2485178573 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1 [Jatropha curcas] |
| 6 |
Hb_000003_500 |
0.2553683563 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 7 |
Hb_053422_010 |
0.2583722053 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 8 |
Hb_002601_050 |
0.2635609242 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 9 |
Hb_004108_180 |
0.2655914663 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 10 |
Hb_003641_030 |
0.2664375839 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 11 |
Hb_008195_060 |
0.2667713172 |
transcription factor |
TF Family: NAC |
NAC transcription factor 018 [Jatropha curcas] |
| 12 |
Hb_000077_170 |
0.2692724178 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 13 |
Hb_001729_030 |
0.2699842381 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 14 |
Hb_001163_110 |
0.2758115237 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 15 |
Hb_028487_060 |
0.2785558236 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 16 |
Hb_000307_040 |
0.2824070917 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 17 |
Hb_001266_150 |
0.2836806896 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 18 |
Hb_001227_070 |
0.284591902 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 19 |
Hb_000105_100 |
0.2850561068 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000866_180 |
0.2901027023 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |