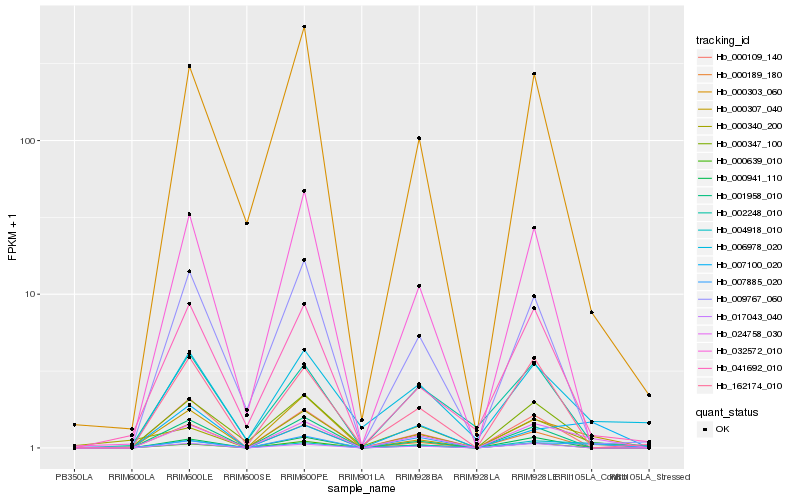
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024758_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 2 |
Hb_004918_010 |
0.1266782416 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000307_040 |
0.1671900573 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 4 |
Hb_000347_100 |
0.2089403895 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 5 |
Hb_017043_040 |
0.2233105586 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_007885_020 |
0.2234158816 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 7 |
Hb_000639_010 |
0.2250721927 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 8 |
Hb_000340_200 |
0.2288797676 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 9 |
Hb_007100_020 |
0.229010636 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 10 |
Hb_006978_020 |
0.2315370048 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000941_110 |
0.2357519419 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 12 |
Hb_032572_010 |
0.2407693478 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 13 |
Hb_001958_010 |
0.2421597707 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 14 |
Hb_041692_010 |
0.2424744798 |
- |
- |
annexin [Manihot esculenta] |
| 15 |
Hb_000189_180 |
0.2429443454 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 16 |
Hb_162174_010 |
0.2431429977 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002248_010 |
0.2449812838 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 18 |
Hb_000303_060 |
0.2455307128 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000109_140 |
0.2461146283 |
- |
- |
hypothetical protein CICLE_v100314482mg, partial [Citrus clementina] |
| 20 |
Hb_009767_060 |
0.246159035 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |