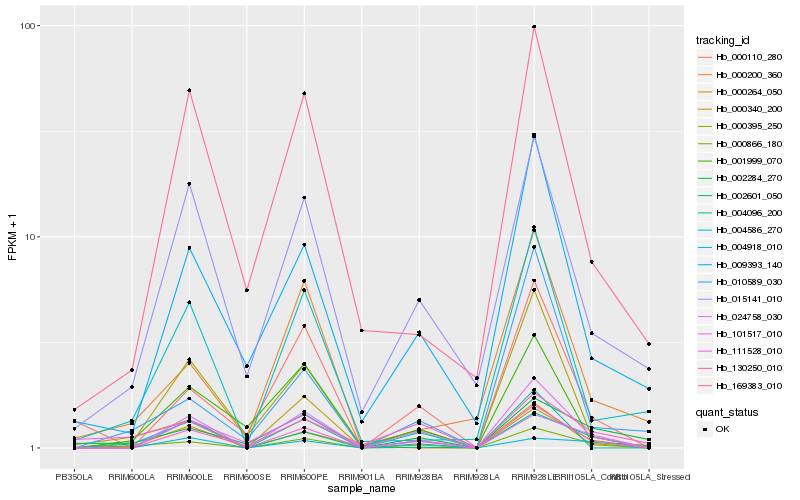
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_180 |
0.0 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 2 |
Hb_101517_010 |
0.2034806569 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 3 |
Hb_111528_010 |
0.2334868418 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 4 |
Hb_002284_270 |
0.2357873171 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_004096_200 |
0.2382796154 |
- |
- |
PREDICTED: GDT1-like protein 5 [Eucalyptus grandis] |
| 6 |
Hb_015141_010 |
0.2418511639 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 7 |
Hb_000340_200 |
0.2468064038 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 8 |
Hb_000110_280 |
0.2481567908 |
- |
- |
PREDICTED: uncharacterized protein LOC105642012, partial [Jatropha curcas] |
| 9 |
Hb_000200_360 |
0.2511717177 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 10 |
Hb_009393_140 |
0.2536306659 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000395_250 |
0.2545834007 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 12 |
Hb_130250_010 |
0.2550213251 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 13 |
Hb_001999_070 |
0.2561910983 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 14 |
Hb_024758_030 |
0.2589726999 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 15 |
Hb_000264_050 |
0.261627766 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 16 |
Hb_002601_050 |
0.2642440884 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 17 |
Hb_004586_270 |
0.2675939254 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_169383_010 |
0.268081765 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004918_010 |
0.2694295514 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_010589_030 |
0.2694925895 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |