| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
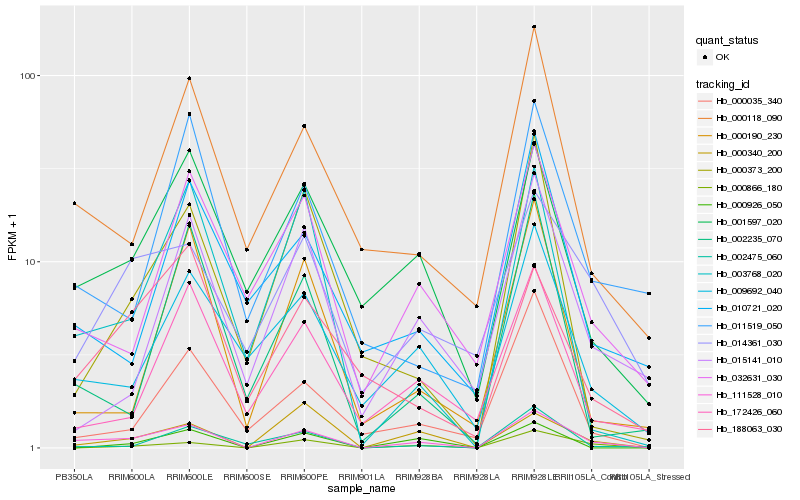
Hb_111528_010 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 2 |
Hb_000866_180 |
0.2334868418 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 3 |
Hb_000118_090 |
0.2379779091 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_009692_040 |
0.2379903774 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001597_020 |
0.2380087675 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003768_020 |
0.2381204957 |
- |
- |
PREDICTED: uncharacterized protein LOC105644157 [Jatropha curcas] |
| 7 |
Hb_000340_200 |
0.2435653905 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 8 |
Hb_032631_030 |
0.2491374895 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 9 |
Hb_014361_030 |
0.2549574245 |
- |
- |
hypothetical protein POPTR_0005s18860g [Populus trichocarpa] |
| 10 |
Hb_000035_340 |
0.2625745286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_188063_030 |
0.2651159375 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 12 |
Hb_172426_060 |
0.2652548198 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 13 |
Hb_000190_230 |
0.2653667376 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 14 |
Hb_011519_050 |
0.2663762426 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 15 |
Hb_002475_060 |
0.2667027155 |
- |
- |
PREDICTED: potassium channel SKOR-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002235_070 |
0.2672481861 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 17 |
Hb_000373_200 |
0.268239815 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 18 |
Hb_000926_050 |
0.2707826818 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 19 |
Hb_010721_020 |
0.2715446599 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_015141_010 |
0.2725163049 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |