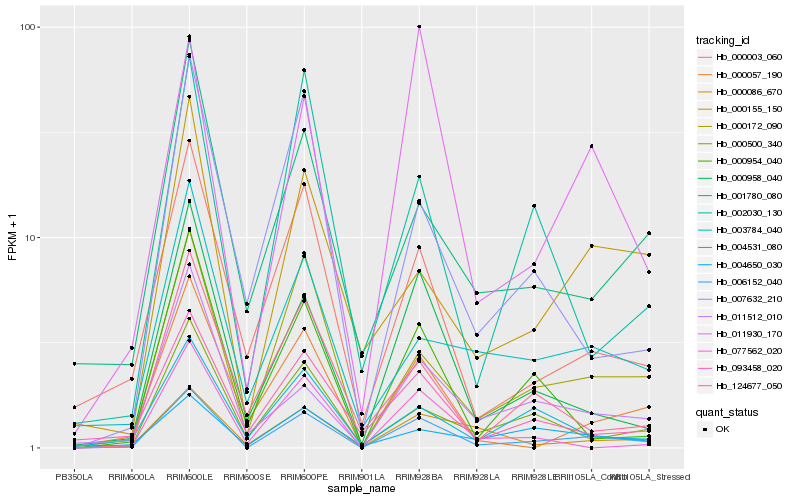
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006152_040 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000172_090 |
0.1653697293 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 3 |
Hb_000958_040 |
0.1773793172 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 4 |
Hb_011930_170 |
0.1917076102 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_000003_060 |
0.1951649899 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 6 |
Hb_093458_020 |
0.1976045746 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 7 |
Hb_000500_340 |
0.2053602062 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 8 |
Hb_000057_190 |
0.2084237645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000954_040 |
0.2104429414 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 10 |
Hb_004650_030 |
0.2111862438 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000155_150 |
0.2127107516 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 12 |
Hb_077562_020 |
0.2134435686 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 13 |
Hb_004531_080 |
0.2138202247 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 14 |
Hb_011512_010 |
0.2182156346 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 15 |
Hb_000086_670 |
0.2187548553 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 16 |
Hb_003784_040 |
0.2195304586 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 17 |
Hb_002030_130 |
0.2227189467 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 18 |
Hb_001780_080 |
0.2227420478 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 19 |
Hb_007632_210 |
0.2240148401 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 20 |
Hb_124677_050 |
0.2246615462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |