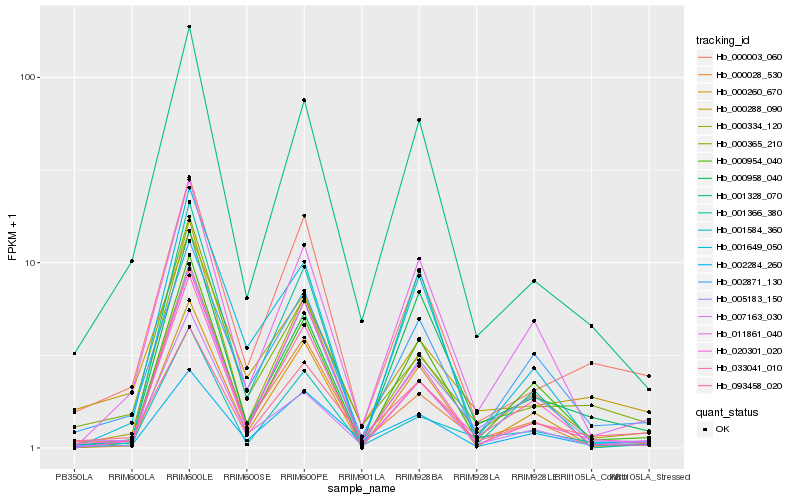
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001328_070 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 2 |
Hb_007163_030 |
0.1290192909 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 3 |
Hb_011861_040 |
0.1345581055 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 4 |
Hb_093458_020 |
0.1350927609 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 5 |
Hb_000958_040 |
0.1393350383 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 6 |
Hb_000003_060 |
0.1413234775 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 7 |
Hb_000365_210 |
0.1438322718 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 8 |
Hb_000334_120 |
0.1458944765 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 9 |
Hb_002284_260 |
0.1470944668 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 10 |
Hb_020301_020 |
0.153098799 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_001649_050 |
0.1562578071 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 12 |
Hb_001584_360 |
0.1565486547 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 13 |
Hb_001366_380 |
0.158327232 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_033041_010 |
0.1605177198 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 15 |
Hb_002871_130 |
0.1609934933 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 16 |
Hb_000954_040 |
0.163009567 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 17 |
Hb_000260_670 |
0.1638949075 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 18 |
Hb_000028_530 |
0.1645664551 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 19 |
Hb_005183_150 |
0.1657801379 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 20 |
Hb_000288_090 |
0.1660860319 |
- |
- |
unnamed protein product [Vitis vinifera] |