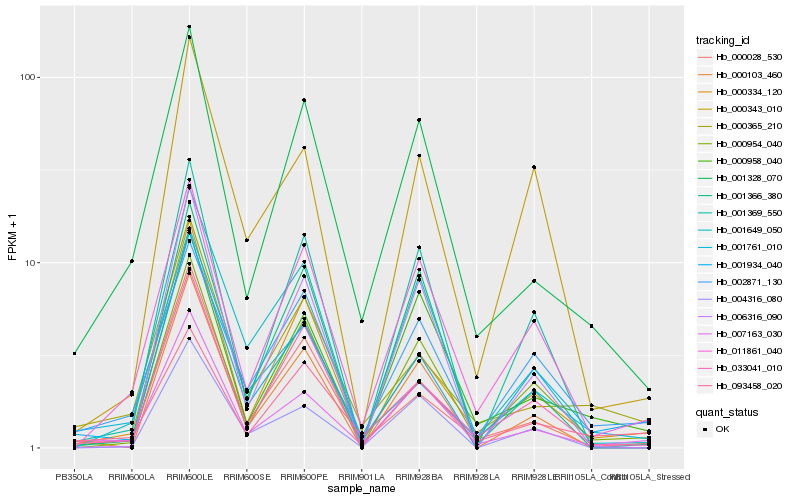
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007163_030 |
0.0 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 2 |
Hb_001328_070 |
0.1290192909 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 3 |
Hb_000958_040 |
0.130791909 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 4 |
Hb_001934_040 |
0.1425087131 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 5 |
Hb_011861_040 |
0.1477935596 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 6 |
Hb_001649_050 |
0.1489748563 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000334_120 |
0.1510615094 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 8 |
Hb_004316_080 |
0.153974811 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 9 |
Hb_000954_040 |
0.1565487891 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 10 |
Hb_000103_460 |
0.158274199 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 11 |
Hb_000365_210 |
0.1623280662 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 12 |
Hb_006316_090 |
0.1632511944 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_000028_530 |
0.1642373287 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 14 |
Hb_002871_130 |
0.1654190846 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 15 |
Hb_001366_380 |
0.1663981163 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001761_010 |
0.1689281503 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 17 |
Hb_093458_020 |
0.17175432 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 18 |
Hb_000343_010 |
0.1724487473 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 19 |
Hb_001369_550 |
0.1767628686 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 20 |
Hb_033041_010 |
0.1770072771 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |