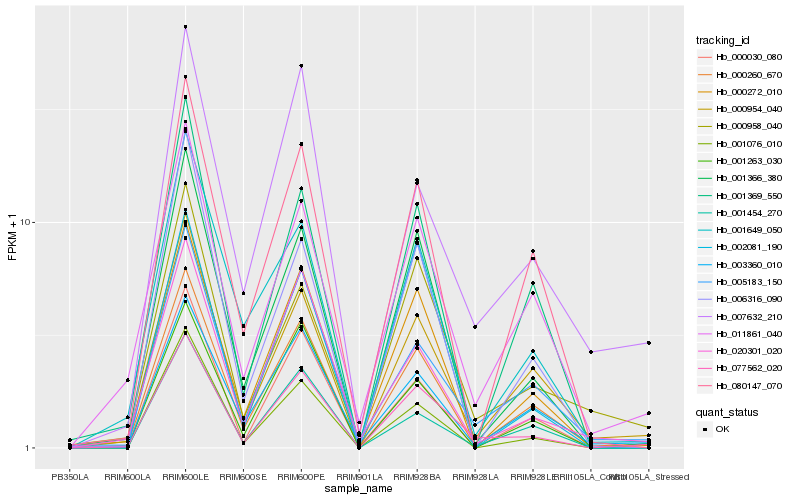
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077562_020 |
0.0 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 2 |
Hb_001366_380 |
0.1232146722 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000260_670 |
0.123919635 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 4 |
Hb_000272_010 |
0.1328669369 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 5 |
Hb_005183_150 |
0.1351932584 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 6 |
Hb_000954_040 |
0.1354921145 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 7 |
Hb_000958_040 |
0.1463968927 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 8 |
Hb_011861_040 |
0.1484245863 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 9 |
Hb_020301_020 |
0.155034079 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_001076_010 |
0.1555064501 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_007632_210 |
0.159292163 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 12 |
Hb_000030_080 |
0.161079551 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 13 |
Hb_003360_010 |
0.1620897026 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 14 |
Hb_001263_030 |
0.1661368323 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 15 |
Hb_001369_550 |
0.1672232164 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 16 |
Hb_001649_050 |
0.1674145127 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 17 |
Hb_002081_190 |
0.1677667069 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 18 |
Hb_006316_090 |
0.1688013314 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_001454_270 |
0.1696198314 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_080147_070 |
0.1705497514 |
- |
- |
Rop5 [Hevea brasiliensis] |